Splicing-based regulation of host responses to cell stress during a bacterial infection
In response to injuring assaults such as infectious challenges, changes in gene expression orchestrate the host responses to damaging threats. Our team investigates how these responses are regulated and coordinated in the context of the infection of human intestinal cells by a foodborne pathogen, Listeria monocytogenes.
This talk will highlight the regulation by alternative splicing of a human stress-sensitive regulator of mRNA stability and translation, cold-inducible RNA-binding protein (CIRBP), in response to membrane damage by bacterial pore-forming toxins, and its role in the control of infection.
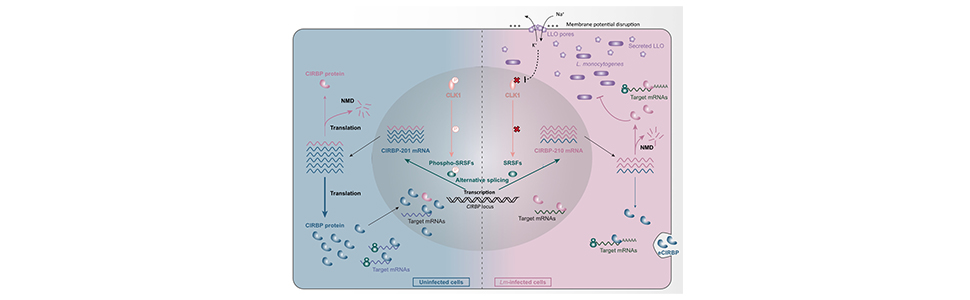
By exploring changes in the splicing landscape of human cells infected by Listeria, we observed that alternative splicing altered the expression of cellular genes involved in host mRNA processing, stability and translation regulation, including SR splicing factors and CIRBP. The pore-forming activity of bacterial secreted toxins was sufficient to drive CIRBP splicing towards unstable transcripts due to the inclusion of NMD-sensitive “poison exons.”
Of note, the expression of the canonical versus the stress-induced CIRBP isoforms had an opposite effect on the ability of cells to control infection. Consistently, in infected cells a larger fraction of CIRBP-bound mRNA targets encoded proteins involved in cell responses to stress and infection than in naive cells. Infection also correlated with the stabilisation or translational repression of some CIRPB targets.
Our study generalises the key contribution of CIRBP in coordinating the responses of cells that face damaging conditions, by extending its relevance to the context of a bacterial infection and of the stress induced by the activity of pore-forming toxins.
By exploring changes in the splicing landscape of human cells infected by Listeria, we observed that alternative splicing altered the expression of cellular genes involved in host mRNA processing, stability and translation regulation, including SR splicing factors and CIRBP. The pore-forming activity of bacterial secreted toxins was sufficient to drive CIRBP splicing towards unstable transcripts due to the inclusion of NMD-sensitive “poison exons.”
Of note, the expression of the canonical versus the stress-induced CIRBP isoforms had an opposite effect on the ability of cells to control infection. Consistently, in infected cells a larger fraction of CIRBP-bound mRNA targets encoded proteins involved in cell responses to stress and infection than in naive cells. Infection also correlated with the stabilisation or translational repression of some CIRPB targets.
Our study generalises the key contribution of CIRBP in coordinating the responses of cells that face damaging conditions, by extending its relevance to the context of a bacterial infection and of the stress induced by the activity of pore-forming toxins.
Mandatory registration for EXTERNAL persons ONLY by clicking here (access badge)
Conférencier(ère)s
Dr. Alice Lebreton
Institut de Biologie de l’École Normale Supérieure
Équipe Dynamique du dialogue bactérie-cellule
Paris
France
