FateCompass: to identify and predict the decisive criteria of cell fate
Cell differentiation is a process regulated by gene expression through the action of transcription factors. Different transcription factors and changes in the order of gene activation lead to different cell fates. In a study published in Cell Reports Methods, the teams of Nacho Molina and Gérard Gradwohl have developed FateCompass, a tool capable of identifying and predicting the impact of these differences on cell specification.
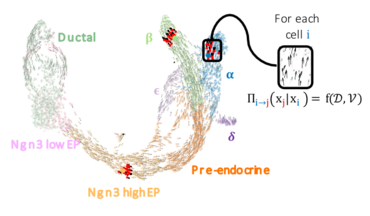
During the process of cell differentiation, genes are switched on and off at precise times by transcription factors, resulting in cell diversity. Understanding the criteria that determine cell fate can shed light on many areas, such as embryogenesis, regenerative medicine, gene and cell therapies, cancer... Currently, to determine the factors impacting cell fate, the expression levels of transcription factors and their target genes are compared. However, it is difficult to accurately measure the dynamic regulatory activity of transcription factors at the level of individual cells and at high throughput.
In this study, the scientists present their FateCompass tool, an approach that exploits single-cell transcriptomic data to identify cell-lineage-specific transcription factors during the differentiation process. To develop it, the researchers used transcriptomic datasets derived from islet cells forming during pancreatic development in mice, or derived, in vitro, from human stem cells. The tool estimates the probability of transition between different cellular states, combining a probabilistic approach with mRNA maturation rate or differentiation potential.By considering dynamic changes and correlations between gene expression and transcription factor activities, FateCompass effectively identifies regulators that are specific to particular lineages. In this way, the tool was able to correctly identify specific regulators already known to be involved in the formation of insulin-secreting ß-cells, and revealed the existence of potential new regulators.
With FateCompass, this study provides a valuable new tool for generating hypotheses and improving understanding of the gene regulatory networks that determine cell fate.

FateCompass is a new tool that helps researchers identify the specific regulators involved in cell differentiation processes. By analyzing transcriptomic data from a single cell, it estimates the dynamic activity of transcription factors and predicts lineage-specific regulators, improving our understanding of the gene regulatory networks that govern cell fate decisions.
Credits: Sacha Jimenez, IGBMC
