Atomic-level structures show how accuracy is maintained in protein synthesis in Eukaryotes
Translation of the genetic code into proteins is realized by repetition of synchronous movements of messenger RNA and transfer RNA through the ribosome. This is the process of translocation. In this article published in the journal Nature, Gulnara Yusupova and Marat Yusupov's team highlights the intermediate steps in translocation carried out by the ribosome in eukaryotic cells, and shows how it manages translocation by preventing frameshifting.
During protein synthesis, messenger RNA (mRNA) and transfer RNA (tRNA) must be rapidly translocated through the ribosome to advance the translational reading frame by one codon. In eukaryotes, translocation is accelerated by elongation factor 2 (eEF2, also known as translocase). Interruption in the maintenance of the mRNA reading frame can lead to the production of aberrant proteins that, if not eliminated, trigger deleterious cellular effects. Until recently, the great complexity of the eukaryotic 80S ribosome hampered high-resolution structural studies of eukaryotic translocation, arguably the most complex mechanistic operation in protein synthesis.
In this study, scientists used cryo-electron microscopy to determine ten high-resolution structures of the Saccharomyces cerevisiae eukaryotic ribosome bound to the full translocation module consisting of mRNA, peptidyl-tRNA and deacylated tRNA. Of the ten intermediates, whose highest resolution reached 1.97 Å, seven were co-resolved with the native Saccharomyces cerevisiae ribosome-bound "translocase" (eEF2). Such a collection of translocation intermediates trapped in the transition from the early to the late state of translocation shows important large-scale conformational changes and reveals how the module composed of mRNA, tRNAs and the nascent peptide chain is moved over distances of around 30 to 50 Å before reaching the near-final translocation position. They have characterized eukaryotic translocation intermediates whose existence had only been hypothesized on the basis of bacterial translocation, but whose reconstructions had never been reported. This study also reveals that the added complexity of the eukaryotic translational apparatus, compared with the bacterial system, is reflected by more sophisticated and finely regulated mechanisms for maintaining the mRNA-tRNA2 module during its translocation through the ribosome.
By shedding light on the intermediate stages of translocation, scientists are helping to refine our understanding of the protein synthesis process in eukaryotic organisms.

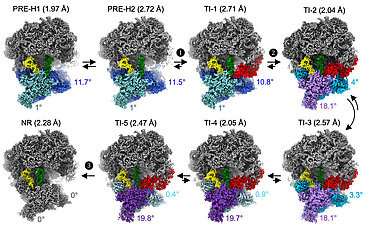
Figure Legend | Translocation of the eukaryotic ribosome.
High-resolution cryo-electron microscopy reconstructions of intermediate translocation states, showing recruitment (1), accommodation (2) and release (3) of elongation factor 2 (eEF2, "translocase", in red) by the eukaryotic ribosome. Black arrows indicate advancement of translocation and possible reversibility of steps. The angles indicate the rotation of the "body" (blue) and "head" (cyan) domains of the small ribosomal subunit relative to the classical (NR) state, around the vertical and perpendicular axes, respectively. After recruitment, translocase accelerates tRNA displacement through five translocation intermediate (TI) states; this displacement is associated with movements of the small ribosomal subunit, namely backward rotation of the "body" domain (blue to grey), forward pivoting of the "head" domain (light blue to violet) and backward pivoting (violet to grey). Finally, the NR intermediate displays the final translocation positions. Two further translocation intermediates (not shown) have been resolved.
