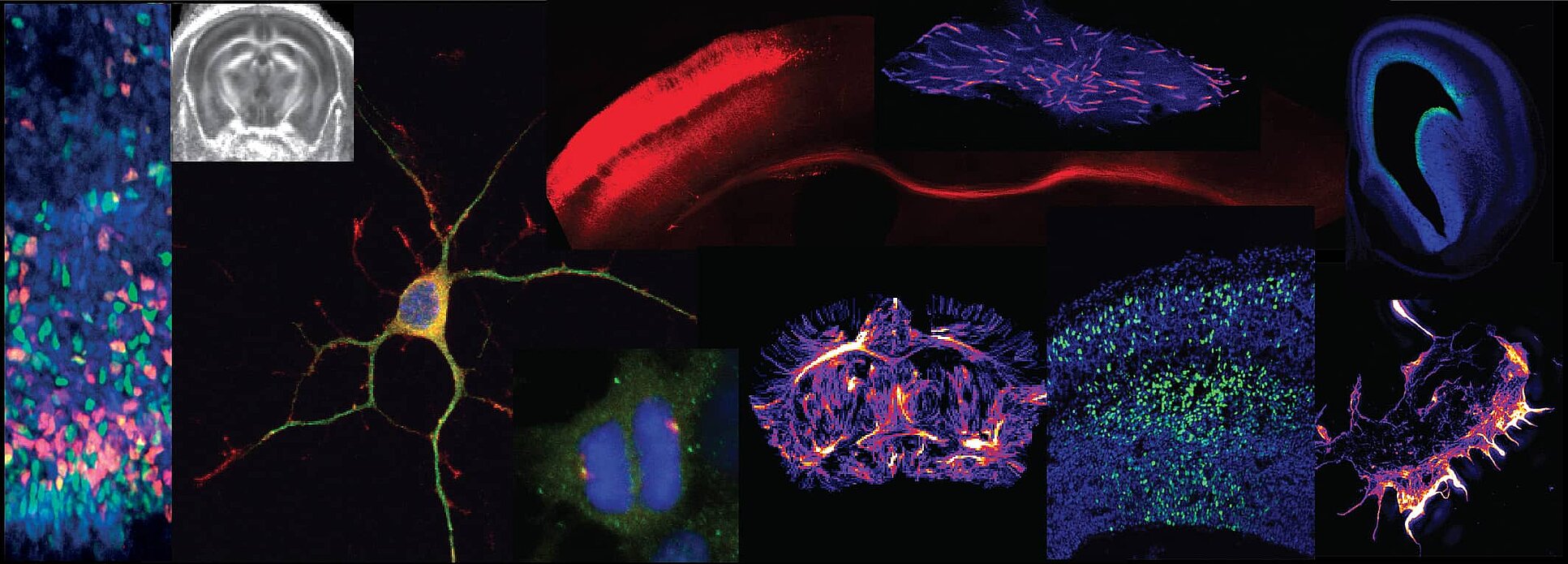
Regulation of cortical development in health and disease
Regulation of cortical development in health and disease
Our lab aims to elucidate the fundamental mechanisms that dictate cell fate acquisition and neuronal maturation during mammalian corticogenesis.
Main interests
Understand the origin of neuronal diversity in the mouse developing cortex
Study post-transcriptional mechanisms that regulate gene expression during cerebral cortex neurogenesis
Investigate the role of cytoskeleton proteins during cortical development
Interpret the pathological mechanisms of associated neurodevelopmental disorders and SUDEP (SUDEP Sudden Unexpected Death in Epilepsy)
Understand the sensitivity of the brain to tRNAs defects.
Strategy
To unravel new regulators of neurodevelopment, we learn from the genetic mutations that have been associated to neurodevelopmental disorders (NDDs), including malformations of cortical development, intellectually disability and epilepsy. Although the etiologies of those diseases are not always clearly understood, the nature of the mutated genes can give us pathological insights and may elucidate general principles for brain development.

Current projects
1. Translational Control of Neuronal Fate and Identity
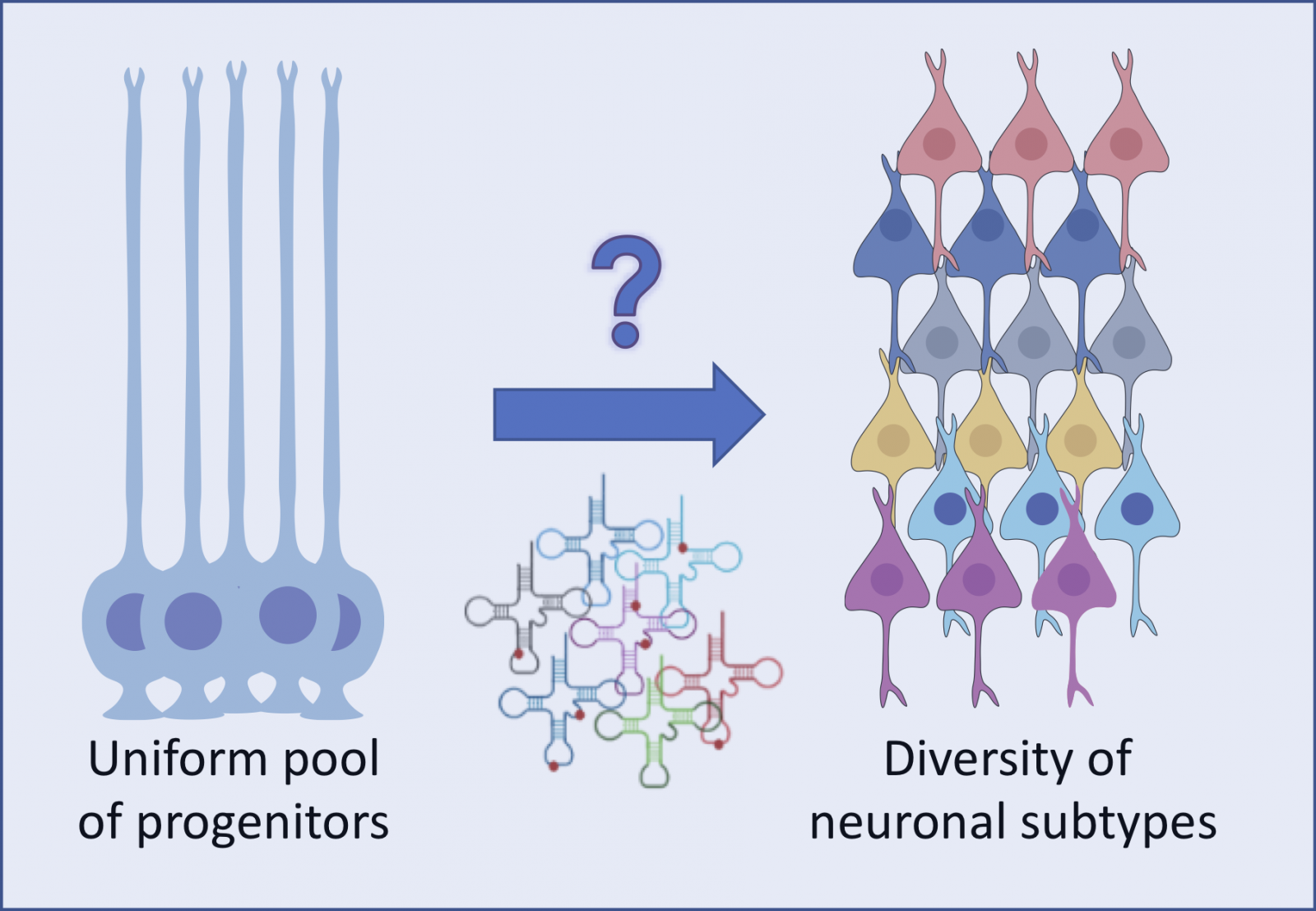 The cerebral cortex is a central structure of the mammalian brain, characterized by a remarkable diversity of neuronal types. Understanding the origin of the extraordinary neuronal diversity is fundamental to understanding how the cortical architecture and its diverse functions emerge during development. It remains a critical challenge in cellular and molecular neurobiology. While the efforts have been focused on transcriptional control, evidence for regulation at the translational level is emerging. We hypothesize that translational control, though oft overlooked, acts in a combinatorial fashion with transcriptional induction to regulate gene expression programs during cortical patterning. We have demonstrated an intimate functional link between cortical development and cellular content of mature transfer RNA (tRNAs) (Laguesse et al, Dev cell, 2015, Del-Pozo-Rodriguez, Brain, 2025), the major determinant of translation. We, therefore, propose to address how translational control, through the modulation of the availability of mature translationally competent tRNAs, hones gene expression programs during lineage progression, thereby regulating neuronal diversity. Studying this yet unexplored question could unravel a hitherto unrecognized level of neuronal fate identity determination in the cerebral cortex
The cerebral cortex is a central structure of the mammalian brain, characterized by a remarkable diversity of neuronal types. Understanding the origin of the extraordinary neuronal diversity is fundamental to understanding how the cortical architecture and its diverse functions emerge during development. It remains a critical challenge in cellular and molecular neurobiology. While the efforts have been focused on transcriptional control, evidence for regulation at the translational level is emerging. We hypothesize that translational control, though oft overlooked, acts in a combinatorial fashion with transcriptional induction to regulate gene expression programs during cortical patterning. We have demonstrated an intimate functional link between cortical development and cellular content of mature transfer RNA (tRNAs) (Laguesse et al, Dev cell, 2015, Del-Pozo-Rodriguez, Brain, 2025), the major determinant of translation. We, therefore, propose to address how translational control, through the modulation of the availability of mature translationally competent tRNAs, hones gene expression programs during lineage progression, thereby regulating neuronal diversity. Studying this yet unexplored question could unravel a hitherto unrecognized level of neuronal fate identity determination in the cerebral cortex
We are combining ribosome profiling, mRNA and tRNA deep sequencing and gene manipulation in vivo in the mouse embryonic cortex to i) uncover the specific translational programs that influence neuronal lineages progression; ii) determine how tRNA repertoires (both at the transcriptional and post-transcriptional levels; i.e. tRNAs modification) are shaped to meet the specific translational needs of different cell types during corticogenesis and iii) validate the functional importance of fluctuation in mature tRNA content during cortical development.
2. Regulation of the intracellular dynamics during cortical development
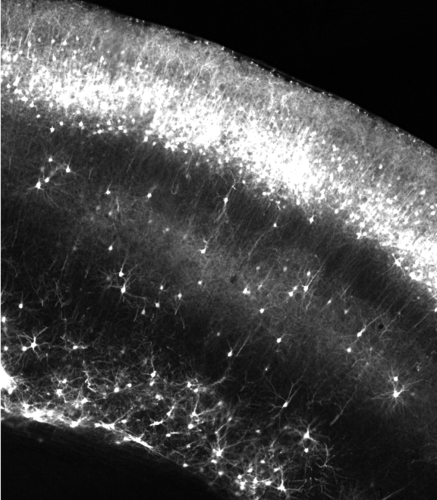
The formation of the nervous system requires functional Microtubules (MTs) and Actin cytoskeletonsduring all stages of development. MTs and actin act in concert with MTs associated proteins (MAPs) and motors and actin binding proteins, respectively, to carry out the structural changes that underpin key developmental events. These include such as neurogenesis, neuronal migration, axon path finding and synapse formation. Interestingly mutations in human genes encoding either tubulin, MAPs or motors have been associated to malformation of cortical development (MCD), a group of socially-devastating neurodevelopmental disorders that can lead to severe intellectual disabilities (ID). However, how these mutations lead to MCD is not fully understood. A better understanding of the role of cytoskeleton proteins during cortical development is critical to shed more light on molecular mechanisms underlying neurodevelopmental pathologies. We approach those issues through a candidates-based approach. Thanks to collaboration with clinicians, we select cytoskeletons associated genes in which variants have been identified in patients presenting with MCD or other neurodevelopmental disorders (NDDs) and whose function is not known in the developing cortex.
We study both the physiological roles of those genes with the aim to identify new regulators of the cortical development and the pathological mechanisms associated to disease to better understand the etiology of NDDs. Our deep functional analysis should allow us to uncover both the cellular and molecular mechanismsunderlying the physiological and pathological function of those newly-discovered genes. We are currently focusing on two genes encoding a kinesin, KIF21b (Asselin et al, Nat com, 2020) (Rivera Alvarez et al, Cell report, 2023) and a MT-associated protein WDR47 (Kannan et al, PNAS, 2017) (Bayam et al, Embo Molecular Medicine, 2024).
3. Regulation of RNA binding proteins (RBPs) during cortical development
RBMX is a gene located on chromosome (chr) X in mammals that encodes the RNA-binding RMBX/hnRNP-G protein, known to regulate alternative splicing as part of the spliceosome machinery. Pathogenic variants in RBMX are associated with a severe but clinically variable neurodevelopmental disorder characterized by intellectual disability, brain and eye malformations affecting hemizygous males. Remarkably, RBMX has many copies in mammalian genomes, including several intronless retrocopies located on autosomes. The most recent retrocopies, RBMXL1 encode likely functional proteins, 96-98% identical to RBMX/Rbmx, expressed during brain development in both species. The main aims of this project are to 1) investigate the mechanisms by which novel human mutations identified in four unrelated families lead to brain disorders and 2) dissect the molecular mechanisms by which RBMX and its functional retrocopy contribute to brain development in both human and mouse. For this purpose, we generated analogous, complementary cellular human and in vivo mouse models and apply multiscale genome-wide approaches and fine-grained lineage tracing in human neural progenitors and mouse cortices. Overall, this project has the potential to unravel how independent, recurrent retrotransposition of a single gene might have contributed to shape brain size and function during evolution.
4. Neurodevelopment basis of Sudden Infant Death Syndrome and Sudden Unexpected Death in Epilepsy(Lead Audrey Farrugia-Jacamon)
Sudden Unexpected Death in Epilepsy (SUDEP) is the most tragic consequence of epilepsy. It is estimated that approximately 1.2 out of every 1,000 people with epilepsy die from SUDEP each year. SUDEP is defined as a sudden, unexpected, non-traumatic, and non-drowning death in people with epilepsy, occurring with or without evidence of a seizure, and where post-mortem investigations—including autopsy, toxicology, and histology—fail to identify a cause of death. Sudden Infant Death Syndrome (SIDS) refers to the sudden, unexplained death of an infant under two years of age (legal definition in France), which remains unexplained after the recommanded post-mortem investigations.We conducted a retrospective genetic analysis of SIDS and SUDEP cases autopsied at the Forensic Institute of Strasbourg. This included 1) a targeted panel of 180 genes implicated in cardiomyopathies, channelopathies, and epilepsy, and 2) solo whole-exome sequencing (WES) when panel analysis failed to detect pathogenic or likely pathogenic variants. Variants of uncertain significance (VUS) were identified in 60% of the cases. Several of these VUS were located in genes involved in neurodevelopment. We now aim to functionally characterize these variants using mouse models, with the goal of better understanding the potential pathophysiological roles of these genes in SIDS and SUDEP.
Members
Researchers
Post-doctoral fellows
PhD students
Engineers
Technicians
Former members
2024: Ekaterina Stoilova - Master 1 Student ; University of Strasbourg
2024: Cécile Roméo - Master 1 Student ; University of Strasbourg
2024: Clémentine Placet - Master 2 Student ; University of Strasbourg
2024: Medhi Krakovic - Master 1 Student ; University of Strasbourg
2018-2023: José RIVERA ALVAREZ - PhD Student (now postdoc at King's College London)
2023: Ikram RMILI NAZIL - 2nd year Bachelor Student ; University of Strasbourg
2022: Juliette HOFFMAN - 3rd year Bachelor Student ; University of Strasbourg
2022: Daniil LUTSKIY - Bachelor Student, King's College London
2022: Ismael VELEZ - 1st year BTS Student ; University of Strasbourg
2018-2021: Hakima FLICI TEZKRATT – PostDoc FRM
2021: Claire RICHARD – Master 1 Student ; University of Strasbourg
2020: Romain CESBRON – Master 1 Student ; University of Strasbourg
2019: Oktay CAKIL – IGBMC Master 1 Summer intern
2019: Karla SORIA ZAVALA – France-Mexico cooperation program - Master 1 intern
2016-2020: Jordi DEL POZO RODRIGUEZ – PhD Student (now Healthcare, Spain)
2015-2019: Laure ASSELIN – PhD Student (now Kaly-Cell)
2017: Théa BEGEL – 2nd year BTS Student ; University of Strasbourg
2017: Mourat VEZIR – IGBMC Master 1 Summer intern
2017: Eloise COLNOT – Master 1 Student ; University Claude Bernard – Lyon
2016: Noémie LELIEVRE – Master 1 Student ; University of Strasbourg
2016: Viviane NGUEFACK NGOUNE – Master 1 Student ; University of Strasbourg
2016: Arthur RADOUX – 2nd year Student ; University of Strasbourg
2015: Morgane FONTAINE – Master 2 Student ; University of Strasbourg
News

Understanding gene regulation during cortical development
At the start of 2022, Juliette Godin, Inserm researcher, won an ERC Consolidator grant for her project "Gene regulation during cortical development".…
Read more
Collaborations and networks
Member of the STRAS&ND Network : Strasbourg Translational Research on the Autism Spectrum Disorder & Neurodevelopmental Disorders
Member of the French Scientific Interest Group on neurodevelopmental disorders and autism
Member of “International Research consortium for the Corpus callosum and cerebral connectivity”.
Member of RISE, Network of all the imaging platforms of Strasbourg
Member, Research Network “DRN club” (development of neuronal circuits) - FR http://www.ibdm.univ-mrs.fr/drn/index.php?page=board
Funding and partners



.png)








2025 Association La Loco de Matteo (to Audrey Farrugia-Jacamon)
2024 ANR PRC TRAP-HD
2024 ANR PRC MultiWW
2024 Association Hélène de Coeur (to Audrey Farrugia-Jacamon)
2024 Post-doctoral fellowship to E. Brivio from Marie Sklodowska-Curie Action
2024 4th year PhD fellowship (P. Tilliole) - FRM (Fondation pour la Recherche Médicale)
2023 ANR PRC pAGOde
2023 Association Hélène de Coeur (to Audrey Farrugia-Jacamon)
2023 Post-doctoral fellowship from FRM to Elena Brivio (Fondation pour la Recherche Médicale)
2023 PhD fellowship (R. Lecat), Ministère de l'Education Nationale, de l'Enseignement supérieur et de la Recherche
2022 ANR PRCI DFG RETRO-RBMX
2022 ERC Consolidator grant
2022 Fondation pour la recherche sur le cerveau- Rotary Espoir en tête
2022 ANR PRC Synovar
2022 4th year PhD fellowship (J. River Alvarez) - FRM (Fondation pour la Recherche Médicale)
2021 PhD fellowship (P. Tilliole), Ministère de l'Education Nationale, de l'Enseignement supérieur et de la Recherche
2021 ANR PRC InotRNA
2019 ANR PRCI TempoCorticoDev
2019 PhD fellowship ( C. Hardion), IDEX, Université de Strasbourg
2018 ANR PRCE DyrkDOWN
2018 ANR PRC NEDD4L-MCD
2016 4th year PhD fellowship (J. Del Pozo) - ARC (Association pour la recherche sur le cancer)
2018 Post-doct. fellowship to H. Flici from FRM (Fondation pour la Recherche Médicale)
2018 Research grant from Fondation Jérome Lejeune
2018 4th year PhD fellowship (L. Asselin) - FRM (Fondation pour la Recherche Médicale)
2016 PhD fellowship (J. Del Pozo) - INSERM/ Region Grand Est
2016 Research grant Idex, Université de Strasbourg
2015 Research grant - Fondation Fyssen
2015 ATIP/Avenir– INSERM
2014 ANR starting grant (young researcher program)
Awards and recognitions
2023 Prix Guy Ourisson (Cercle Gutenberg Alsace)
2020 Prix Espoir University of Strasbourg (https://recherche.unistra.fr/index.php?id=31268)
2019 Prix Wallach - Académie d’Alsace des Sciences, Lettres et Arts
2012 Post-doctoral fellowship to J. Godin from Marie Curie Action – FP7-PEOPLE-2010-IEF
2011 Post-doctoral long term fellowship to J. Godin from EMBO – ALTF-1031-2010
2010 Post-doctoral fellowship from FRM (Fondation pour la Recherche Médicale)
Publications
-
2018
-
Ciliogenesis and cell cycle alterations contribute to KIF2A-related malformations of cortical development
- Loïc Broix
- Laure Asselin
- Carla Silva
- Ekaterina Ivanova
- Peggy Tilly
- Johan Gilet
- Nicolas Lebrun
- Hélène Jagline
- Giuseppe Muraca
- Yoann Saillour
- Nathalie Drouot
- Madeline Louise Reilly
- Fiona Francis
- Alexandre Benmerah
- Nadia Bahi-Buisson
- Richard Belvindrah
- Laurent Nguyen
- Juliette Godin
- Jamel Chelly
- Maria-Victoria Hinckelmann
Human Molecular Genetics ; Volume: 27 ; Page: 224-238
-
-
2017
-
WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy
- Meghna Kannan
- Efil Bayam
- Christel Wagner
- Bruno Rinaldi
- Perrine Kretz
- Peggy Tilly
- Marna Roos
- Lara Mcgillewie
- Séverine Bär
- Shilpi Minocha
- Claire Chevalier
- Chrystelle Po
- Jamel Chelly
- Jean-Louis Mandel
- Renato Borgatti
- Amélie Piton
- Craig Kinnear
- Ben Loos
- David Adams
- Yann Herault
- ...
Proceedings of the National Academy of Sciences of the United States of America ; Volume: 114 ; Page: E9308-E9317
-
-
2016
-
Mutations in the HECT domain of NEDD4L lead to AKT–mTOR pathway deregulation and cause periventricular nodular heterotopia
- Loic Broix
- Hélène Jagline
- Ekaterina L Ivanova
- Stéphane Schmucker
- Nathalie Drouot
- Jill Clayton-Smith
- Alistair T. Pagnamenta
- Kay Metcalfe
- Bertrand Isidor
- Ulrike Walther Louvier
- Annapurna Poduri
- Jenny Taylor
- Peggy Tilly
- Karine Poirier
- Yoann Saillour
- Nicolas Lebrun
- Tristan Stemmelen
- Gabrielle Rudolf
- Giuseppe Muraca
- Benjamin Saintpierre
- ...
Nature Genetics ; Volume: 48 ; Page: 1349-1358
-
Emerging Roles for the Unfolded Protein Response in the Developing Nervous System
- Juliette Godin
- Catherine Creppe
- Sophie Laguesse
- Laurent Nguyen
Trends in Neurosciences ; Volume: 39 ; Page: 394-404
-
Real‐time Recordings of Migrating Cortical Neurons from GFP and Cre Recombinase Expressing Mice
- Sylvia Tielens
- Juliette Godin
- Laurent Nguyen
Current Protocols in Neuroscience ; Volume: 74
-
Page 2 of 2





