Genomic and epigenomic regulation of cell fate
Genomic and epigenomic regulation of cell fate
DNA is packaged in the form of “chromatin” including nucleosomes, epigenetic modifications and a large number of proteins associated with various biological processes. Chromatin regulators allow genetic information to be decoded differently depending on cellular context. In this way, unique gene expression programs are activated in each cell type, leading to distinct phenotypes and specialised functions.
How is cellular identity regulated during developmental processes, or disrupted in pathological conditions? In our laboratory, we aim to address this fundamental question using cutting-edge genetic engineering, biochemical assays and high-throughput sequencing technologies. In particular, our research focuses on transcription factors and epigenetic regulators, which are critical for embryonic development and often de-regulated in human diseases such as cancer and neurodevelopmental disorders.
We take advantage of mouse embryonic stem cells as a model system for cell fate decisions. Using genetically manipulated pluripotent cells, we can assess not only the influence of proteins of interest on the transcriptome and epigenome, but also test their phenotypic impact on cellular differentiation in vitro and embryonic development in vivo.

RECENT PUBLICATIONS
Tetramerisation governs SALL transcription factor function in development and disease. S. Giuliani, K. Chhatbar, M. Wear, J. Guy, T. Mathieson, H. Burdett, L. George, G. Alston, C. Spanos, T. McHugh, D. Kelly, R. Pantier and A. Bird. bioRxiv (preprint), 2025. https://doi.org/10.1101/2025.10.27.684836
TET knockout cells transit between pluripotent states and exhibit precocious germline entry. R. Pantier, E. Barbieri, S. Gonzalez Brito, E. Thomson, T. Tatar, D. Colby, M. Zhang, I. Chambers. The EMBO Journal, 2025. https://doi.org/10.1038/s44318-025-00597-9
MeCP2 binds to methylated DNA independently of phase separation and heterochromatin organisation. R. Pantier, M. Brown, S. Han, K. Paton, S. Meek, T. Montavon, N. Shukeir, T. McHugh, D.A. Kelly, T. Hochepied, C. Libert, T. Jenuwein, T. Burdon, A. Bird. Nature Communications, 2024. https://doi.org/10.1038/s41467-024-47395-1
Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome. J.A. Watson, R. Pantier, U. Jayachandran, K. Chhatbar, B. Alexander-Howden, V. Kruusvee, M. Prendecki, A. Bird, A.G. Cook. Life Science Alliance, 2023. https://doi.org/10.26508/lsa.202201588
High-throughput sequencing SELEX for the determination of DNA-binding protein specificities in vitro. R. Pantier, K. Chhatbar, G. Alston, H.Y. Lee, A. Bird. STAR Protocols, 2022. https://doi.org/10.1016/j.xpro.2022.101490
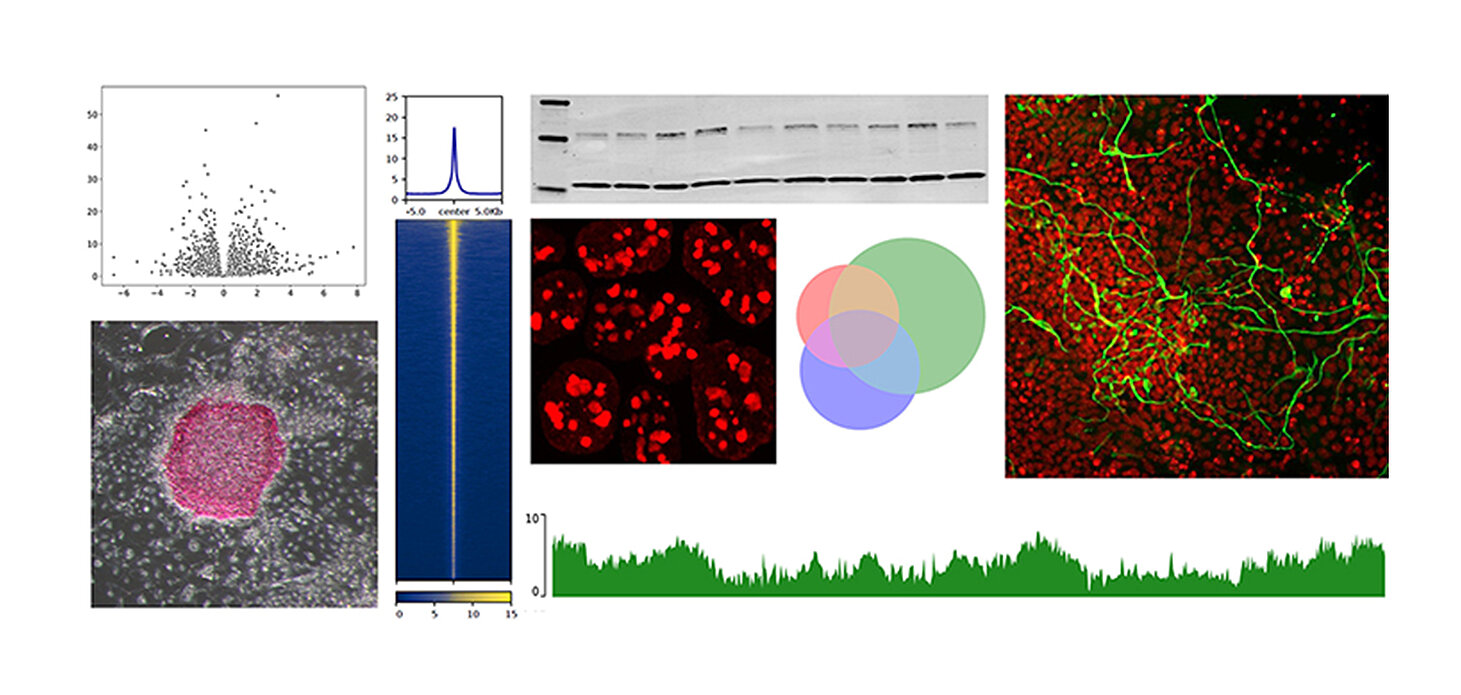
SALL4 controls cell fate in response to DNA base composition. R. Pantier, K. Chhatbar, T. Quante, K. Skourti-Stathaki, J. Cholewa-Waclaw, G. Alston, B. Alexander-Howden, H.Y. Lee, A.G. Cook, C.G. Spruijt, M. Vermeulen, J. Selfridge, A. Bird. Molecular Cell, 2021. https://doi.org/10.1016/j.molcel.2020.11.046
TET1 Interacts Directly with NANOG via Independent Domains Containing Hydrophobic and Aromatic Residues. R. Pantier, N. Mullin, E. Hall-Ponsele, I. Chambers. Journal of Molecular Biology, 2020. https://doi.org/10.1016/j.jmb.2020.10.008
Endogenous epitope-tagging of Tet1, Tet2 and Tet3 identifies TET2 as a naïve pluripotency marker. R. Pantier, T. Tatar, D. Colby, I. Chambers. Life Science Alliance, 2019. https://doi.org/10.26508/lsa.201900516
Members
Researchers
PhD students
Current projects
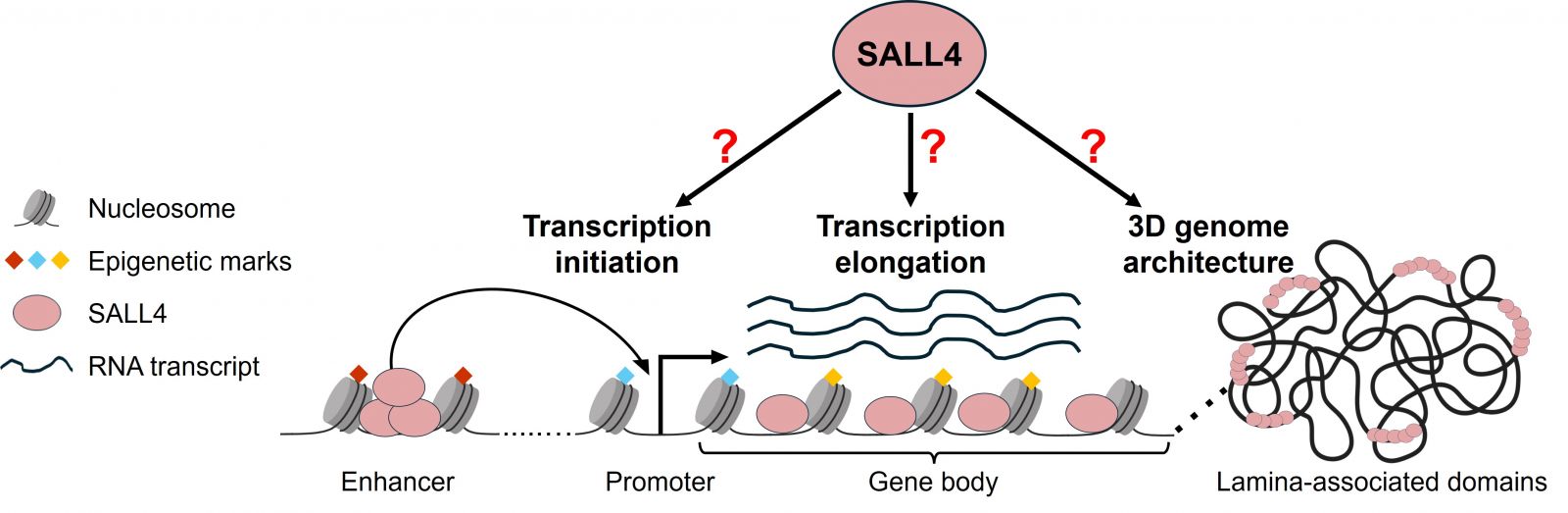
Deciphering the principles of gene regulation by the atypical transcription factor SALL4 (AT-BASE)
Our recent work demonstrated that the stem cell factor SALL4 controls transcription in proportion to the A/T nucleotide content of its target genes. This project aims at uncovering the molecular mechanisms underlying this new mode of gene regulation at three different genomic scales:
- Enhancers: regulation of transcription initiation via long-range looping with promoters of target genes?
- Gene bodies: regulation of transcription elongation via local binding of SALL4 over gene units?
- Lamina-associated domains: regulation of 3D genome architecture via dispersed binding of SALL4 over the AT-rich genome?
Funding and partners
- 2025: Young Researchers program (JCJC) – French National Research Agency (ANR)
- 2025: “Tremplin ERC” funding (T-ERC) – French National Research Agency (ANR)
- 2025: “Attractivity” Excellence initiative (IdEx) – University of Strasbourg
- 2025: Excellence laboratory (LabEx) Startup package – IGBMC





