Cell cycle and ubiquitin signaling
Cell cycle and ubiquitin signaling
Cell cycle is a fundamental biological process that ensures organism development and its dysregulation is linked to many human diseases. Correct execution of cell division requires duplication of all genetic material during S-phase followed by its precise partitioning between two daughter cells during mitosis. During this process, not only the genetic material but also the cellular organelles have to be equally distributed to the newborn cells to allow for their proper functioning. Cell cycle machinery involves many critical factors and enzymes that need to be precisely regulated in time and space. How is their correct and timely localization to the subcellular compartments achieved during different cell cycle transitions, remains an important question in the field.
One of the molecular mechanisms driving fidelity of cell division is ubiquitylation (or ubiquitination), which is a covalent, posttranslational modification of substrates by a small protein ubiquitin. Ubiquitylation frequently targets the substrates to the 26S proteasome, which serves as the main selective system for protein degradation. However, ubiquitylation of substrate proteins can also result in non-proteolytic events and plays important functions in many biological processes including cell proliferation. Our group described the role of non-proteolytic ubiquitin signaling in the regulation of essential kinases during mitosis of human cells. However, little is known on how ubiquitylation of these essential substrates determines their function and how these pathways are dysregulated in human diseases.
Using our expertise in cell biology, we aim at discovering novel essential pathways that regulate equal partitioning of genetic material and cellular organelles during mitosis and re-assembly of nuclear pore complexes after successful completion of cell division. We wish to understand both ubiquitin-dependent and ubiquitin-independent regulatory mechanisms ensuring localization of essential factors involved in these processes. In particular, we focus our current efforts on the ubiquitin-binding proteins (UBDs), which interact with ubiquitylated substrates and mediate their cellular fates and downstream signaling. Finally, we aim at identification of possible links of these pathways to human pathologies such as cancer and neurological diseases.

Members
Researchers
Post-doctoral fellows
PhD students
Former members
- Kyrylo Tron (University of Göttingen, Germany)
- Sadek Fournane
- Stephane Schmucker (University of Strasbourg)
- Paulo Costa (University of Strasbourg)
- Thibaud Metzger (Thermo Fisher Scientific)
- Ksenia Krupina (UCSD, San Diego, USA)
- Katerina Jerabkova (University of Strasbourg)
- Sushil Awal (CRBM, Montpellier, France)
- Arantxa Agote Aran (ETH Zurich, Switzerland)
- Aurore Vuidel (Ksilink, Strasbourg, France)
- Inès Jmel Boyer (University of Dundee, UK)
Current projects
1. The role of the ubiquitin-binding protein UBASH3B in mitosis and cancer
Defects in mitotic pathways lead to aneuploidy and tetraploidy, which are highly prevalent in cancer disease. Deregulated expression of mitotic factors is a common trait of various cancers stimulating development of antimitotic therapeutic targets. Attachment of ubiquitin to substrate proteins regulates proteolytic and non-proteolytic pathways during mitosis. The ubiquitin-binding proteins determine the fate of the ubiquitylated substrates and serve as intracellular “receptors” for the specific ubiquitin signals. The ubiquitin receptors (Ub-receptors) represent a novel and attractive class of druggable targets but to date no inhibitory small molecules exist that could be used in cancer therapies. In collaboration with the High Throughput Cell-based Screening facility at the IGBMC, we have performed a visual, high-throughput siRNA screen using customized, human "Ubiquitin Decoders" library and developed multi-parametric control-based gene selection protocols. The hit list contains many novel mitotic UBDs. One of the UBD proteins, UBASH3B was already characterized in detail in our laboratory.
UBASH3B acts as a receptor for mitotic kinase, Aurora B that is critical for timing and fidelity of chromosome segregation during mitosis. Unlike known Ub-receptors transferring substrates for their proteolytic degradation, UBASH3B does not regulate protein levels of Aurora B. UBASH3B also does not regulate kinase activity of Aurora B. Instead, UBASH3B controls the subcellular distribution of this kinase. UBASH3B localizes to the mitotic spindle and is both required and sufficient to transfer Aurora B from chromosomes to microtubules.
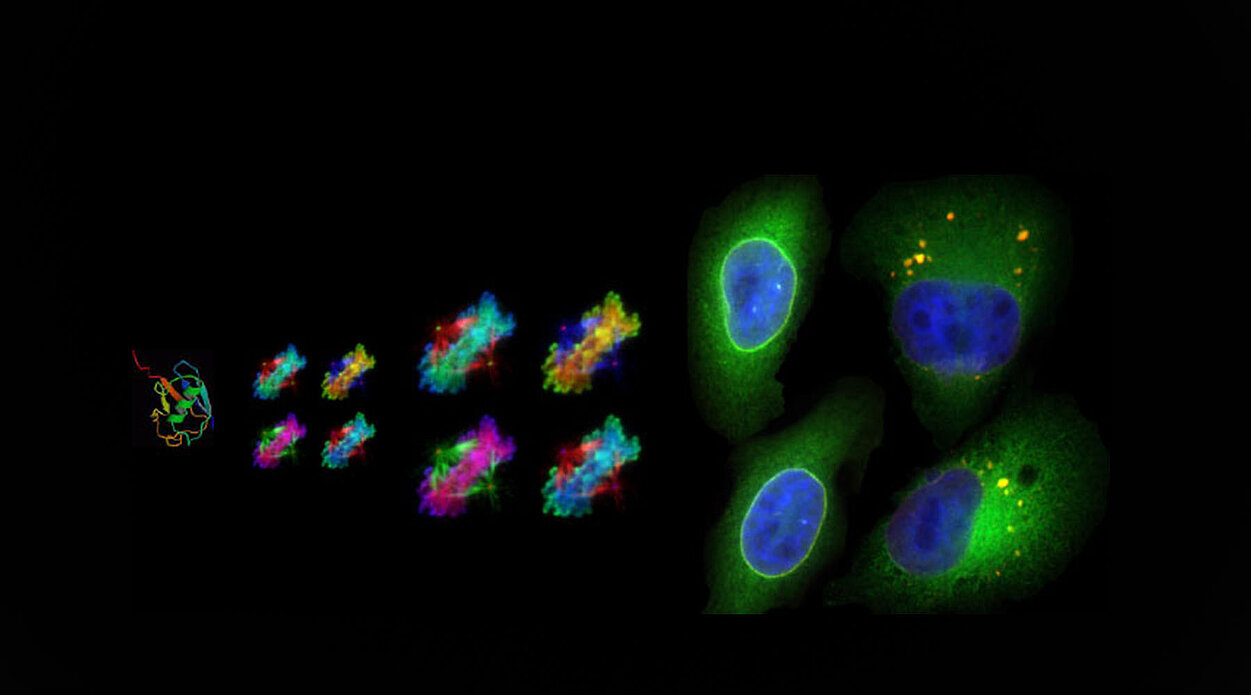
Fig. 1. (A) Model for regulation of localization and function of Aurora B by the ubiquitin receptor UBASH3B. Aurora B (red) is ubiquitylated by CUL3 (light blue) and interacts with microtubule-associated UBASH3B (yellow) prior to anaphase. UBASH3B is required and sufficient to transfer of Aurora B from the centromeric regions of chromosomes (blue) to the spindle microtubules (black lines) thereby controlling its centromeric and kinetochore (grey) as well as spindle functions. UBASH3B acts in concert with MKlp2 (green) targeting Aurora B to the future midzone microtubules and controls timing and fidelity of chromosome segregation. (B) Schematic model depicting a switch-like mode of regulation Aurora B localization by UBASH3B and thereby Spindle assembly checkpoint (SAC) signaling. Low levels of UBASH3B induce chromosomal Aurora B spreading, activation of SAC and persistent mitotic arrest followed by apoptosis. Upregulated levels of UBASH3B induce premature re-localization of Aurora B to microtubules, inhibition of SAC and aneuploidy.
Our data demonstrate that redistribution of Aurora B from centromeres to microtubules by UBASH3B controls timing and fidelity of chromosome segregation and thereby euploidy of cells (Fig. 1).
Our new data also suggest that UBASH3B represents an ideal drug candidate for cancer therapies. First, despite its ubiquitous expression in the body, mice lacking UBASH3B have no detectable phenotypes. Second, its levels were shown to be upregulated in patient samples of highly aggresive cancers. Third, in animal models of breast cancer UBASH3B promotes invasion and metastasis. Finally, our data show that siRNA-mediated downregulation of UBASH3B specifically in aggressive cancer cells leads to their apoptotic death. Thus, UBASH3B represents a novel target, specific for aggresive anauploid cancerous cells. Based on these results, we propose that interfereing with the function of UBASH3B in cancer cells will lead to their apoptotic death but will not dramaticaly influence normally dividing, primary cells. Therefore, we have identified a set of small molecule inhibitors that can specifically bind to and inactivate UBASH3B during mitosis. Treatment with these inhibitors leads to mitotic arrest and cell death specifically in cancer but not in normal primary cells. The goal of this research project is to validate UBASH3B inhibitors as anti-cancer lead compounds for development of a future drug. The long-term perspectives are to expand this strategy to other ubiquitin-based pathways which are critical for mitotic progression.
2. Regulation of assembly of nuclear pore complexes
Nuclear Pore Complexes (NPCs) constitute the sole communication gates between the nucleus and the cytoplasm and ensure the cell’s function and survival. They are inserted in the nuclear envelope (NE), a double membrane structure surrounding the cell nucleus and mediate the transport and exchange of proteins and RNAs between the nucleus and the cytoplasm. Nucleoporins (Nups) are the building protein units of the NPCs. In the cytoplasm, the soluble Nups exist, but how their assembly into NPCs is restricted to the NE remains a mystery. Recently, our team found that the family of the fragile-X related proteins (FXRPs) (FXR1, FXR2 and FMRP) and microtubule-based transport drive spatial assembly of Nups at NE in human cells. In the absence of FXRPs or microtubules, Nups aberrantly self-assemble in the cytoplasm into structures named cytoplasmic nucleoporin granules (CNGs), and protein export and cell cycle progression are perturbed (Fig. 2). Importantly, the models of the Fragile-X syndrome (FXS) where FMRP is absent, display CNGs, suggesting that these defects may contribute to FXS pathology. FXS is a rare genetic neurodevelopmental syndrome and the most frequent familial form of intellectual disability. FXS patients also display a range of developmental defects including inappropriate behaviors, facial malformations and macroorchidism. Currently, no cure exists for FXS. The main objective of this research project is to understand if and how defects in Nups homeostasis mediate FXS pathology. We hypothesize that defects in the Nup pathway may perturb neuronal development through one or several possible mechanisms: inhibition of nucleocytoplasmic transport of specific proteins and/or RNAs or factors required for faithful cell cycle progression due to their sequestration to CNGs and/or disruption of localized translation processes of Nups. This research will likely help our understanding of how misregulation of Nups can contribute to FXS and may create novel therapeutic perspectives for FXS patients.
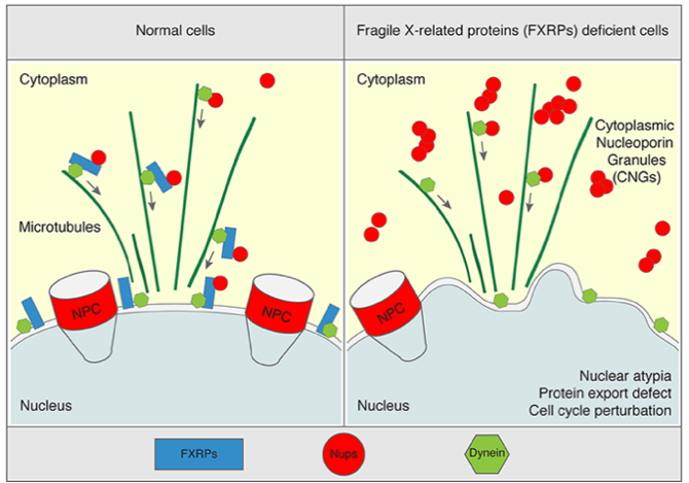
Fig. 2. A model how FXRPs control Nup homeostasis. FXRPs (blue) interact with cytoplasmic soluble Nups (red circles) and dynein (green) and facilitate their localization to the NE during early G1. Silencing of FXRPs or disruption of microtubule-based transport, leads to the accumulation of the CNGs, protein export defects, and defects in cell cycle progression, which may contribute to the pathology of FXS. Adapted from Agote-Aran et al, EMBO J 2020.
3. The role of novel ubiquitin-binding proteins during mitotic exit and assembly of nuclear pore complexes
In search for novel ubiquitin-binding proteins regulating cell division, we identified NICE4/UBAP2L as an important factor for proper execution of mitotic exit. We could demonstrate that NICE4 regulates fidelity of mitotic exit and reformation of the nuclear envelope (NE) at the end of mitosis as inactivation of NICE4 in human cells leads to irregularly shaped polylobbed nuclei and defects in nucleoporin assembly.
Interestingly, mass spectrometry analysis identified the Fragile-X related (FXR) family as interactors of NICE4/UBAP2L, suggesting that NICE4/UBAP2L might be involved in the regulation of the Nups assembly pathway. Indeed, NICE4/UBAP2L can strongly interact with FXR1 and FRMP and CRISPR/Cas9 NICE4/UBAP2L knock-out cells display the same Nups condensation and nuclear defects as FXRPs-deficient cells. NICE4/UBAP2L depletion can also inhibit localization of FXRPs to the nuclear envelope and leads to cytoplasmic aggregation FXRPs, suggesting that NICE4/UBAP2L may spatially regulate their function and act as a chaperone for correct cellular localization of FXRPs. Interestingly, the function of NICE4/UBAP2L as a chaperone for FXRPs is exclusively mediated by its RGG domain located in the N-terminal part of the protein and the RGG domain is sufficient to interact with FXRPs, suggesting that NICE4/UBAP2L regulates FXRPs in an ubiquitin-independent manner. It remains to be studied how exactly NICE4/UBAP2L contributes to the regulation of Nups assembly at the nuclear envelope and to cellular proliferation and how defects in this pathway may contribute to human diseases such as cancer and fragile x syndrome.
Moreover, we aim at identification of targets of NICE4/UBAP2L, which could be recognized in an ubiquitin-dependent manner and may contribute to the regulation of fidelity of mitotic exit. Our long-term future activities will also concentrate on identification of other crucial ubiquitin-binding proteins required for cell cycle progression.
Publications
2024
Pre-publication, Working Document
Annulate Lamellae biogenesis is essential for nuclear pore function
- Junyan Lin
- Arantxa Agote-Arán
- Yongrong Liao
- Rafael Schoch
- Paolo Ronchi
- Victor Cochard
- Rui Zhu
- Charlotte Kleiss
- M. Ruff
- Guillaume Chevreux
- Yannick Schwab
- Bruno P. Klaholz
- Izabela Sumara
Article in a journal
Cytoplasmic nucleoporin assemblage: the cellular artwork in physiology and disease
- Junyan Lin
- Izabela Sumara
Nucleus ; Volume: 15 ; Page: 2387534
Article in a journal
UBAP2L ensures homeostasis of nuclear pore complexes at the intact nuclear envelope
- Yongrong Liao
- Leonid Andronov
- Xiaotian Liu
- Junyan Lin
- Lucile Guerber
- Linjie Lu
- Arantxa Agote-Arán
- Evanthia Pangou
- Li Ran
- Charlotte Kleiss
- Mengdi Qu
- Stephane Schmucker
- Luca Cirillo
- Zhirong Zhang
- Daniel Riveline
- Monica Gotta
- Bruno P Klaholz
- Izabela Sumara
Journal of Cell Biology ; Volume: 223 ; Page: e202310006
2023
Article in a journal
UBAP2L ‐dependent coupling of PLK1 localization and stability during mitosis
- Lucile Guerber
- Aurore Vuidel
- Yongrong Liao
- Charlotte Kleiss
- Erwan Grandgirard
- Izabela Sumara
- Evanthia Pangou
EMBO Reports ; Volume: 24
Article in a journal
CaMK1D signalling in AgRP neurons promotes ghrelin-mediated food intake
- Karl Vivot
- Gergo Meszaros
- Evanthia Pangou
- Zhirong Zhang
- Mengdi Qu
- Eric Erbs
- Gagik Yeghiazaryan
- Mar Quiñones
- Erwan Grandgirard
- Anna Schneider
- Etienne Clauss-Creusot
- Alexandre Charlet
- Maya Faour
- Claire Martin
- Fedor Berditchevski
- Izabela Sumara
- Serge Luquet
- Peter Kloppenburg
- Rubén Nogueiras
- Roméo Ricci
Nature Metabolism ; Volume: 5 ; Page: 1045-1058
Article in a journal
KCNN4 links PIEZO-dependent mechanotransduction to NLRP3 inflammasome activation
- Li Ran
- Tao Ye
- Eric Erbs
- Izabela Sumara
- Romeo Ricci
- Zhirong Zhang
- Stephan Ehl
- Nathalie Spassky
Science Immunology ; Volume: 8 ; Page: adf4699
2022
Article in a journal
Non-proteolytic ubiquitylation in cellular signaling and human disease
- Yongrong Liao
- Izabela Sumara
- Evanthia Pangou
Communications Biology ; Volume: 5 ; Page: 114
Pre-publication, Working Document
UBAP2L-dependent coupling of PLK1 localization and stability during mitosis
- Lucile Guerber
- Evanthia Pangou
- Aurore Vuidel
- Yongrong Liao
- Charlotte Kleiss
- Erwan Grandgirard
- Izabela Sumara
Article in a journal
Phosphorylation of XPD drives its mitotic role independently of its DNA repair and transcription functions
- Emmanuel Compe
- Evanthia Pangou
- Nicolas Le May
- Clémence Elly
- Cathy Braun
- Ji-Hyun Hwang
- Frédéric Coin
- Izabela Sumara
- Kwang-Wook Choi
- Jean-Marc Egly
Science Advances ; Volume: 8
Article in a journal
Ubiquitin Binding Protein 2-Like (UBAP2L): is it so NICE After All?
- Lucile Guerber
- Evanthia Pangou
- Izabela Sumara
Frontiers in Cell and Developmental Biology ; Volume: 10
Collaborations and networks
- Kay Hofmann (University of Cologne, Germany)
- Radislav Sedlacek (University of Prag, Czech Republic)
- Pascal Villa (PCBIS, Illkirch France)
- Dominique Bonnet (University of Strasbourg, France)
- Karim Hnia (I2MC, Toulouse, France)
- Julien Vermot (Imperial College London, London, UK)
- Valerie Doye (IJM, Paris , France)
- Claudia Bagni (University of Lausanne, Switzerland)
- Yannick Schwab (EMBL Heidelberg, Germany)
- Romeo Ricci (IGBMC, Illkirch, France)
- Catherine Tomasetto (IGBMC, Illkirch, France)
- Jamel Chelly (IGBMC, Illkirch, France)
- Hervé Moine (IGBMC, Illkirch, France)
- Jean-Louis Mandel (IGBMC, Illkirch, France)
- Bruno Klaholz (IGBMC, Illkirch, France)
Awards and recognitions
- Prime d’Excellence Scientifique from CNRS, 2011-2014, 2016-2020
- University of Strasbourg Institute for Advanced Study (USIAS) 2014
- Sanofi Innovation Awards Program Europe (iAwards) 2018
Funding and partners
- ATIP-Avenir Research Grant (R10076MS) 2010
- “Programme de Mécénat“ with Sanofi Aventis and Aviesan 2010
- La Ligue Contre le Cancer Grand Est (CCIR-GE) - Project grant 2013, 2014, 2015, 2020
- Fondation ARC pour la recherche sur le cancer - Project grant 2012, 2014
- University of Strasbourg Institute for Advanced Study (USIAS) 2014
- SATT Conectus Alsace 2015, 2019
- Fondation ARC pour la recherche sur le cancer - Programme Labellisé Fondation ARC pour la recherche sur le cancer 2016
- L'Institut National du Cancer (INCa) 2016 - Projets libres de Recherche Biologie et Sciences du Cancer 2016
- Agence Nationale de la Recherche (ANR) AAPG project 2017
- Sanofi Innovation Awards Program Europe (iAwards) 2018
- Programme Fédérateur Aviesan: Vers une nouvelle carte subcellulaire de la cellule cancéreuse, Plan Cancer 2019

