Spatial organisation of the genome
Spatial organisation of the genome
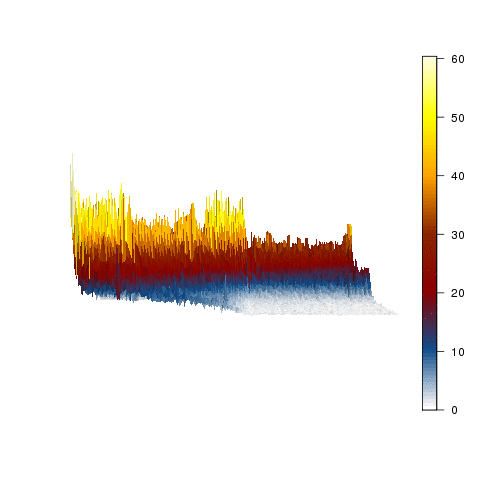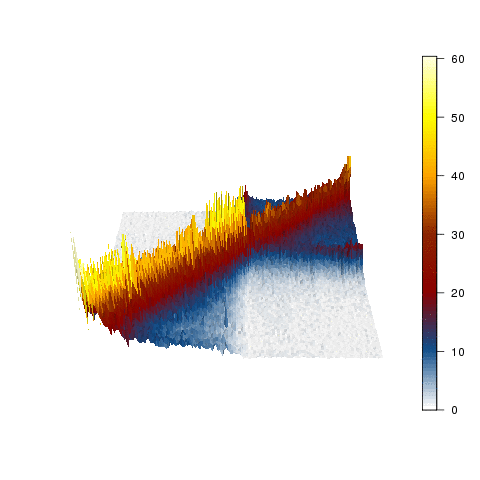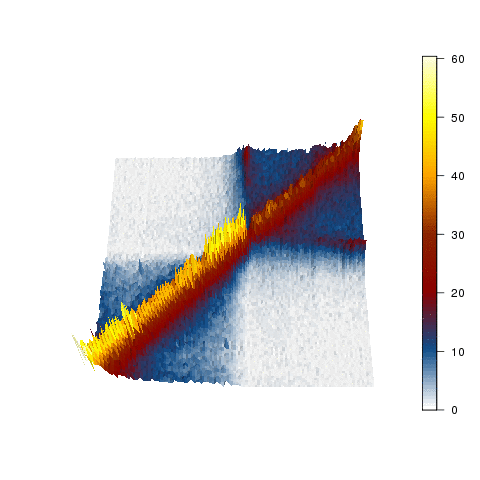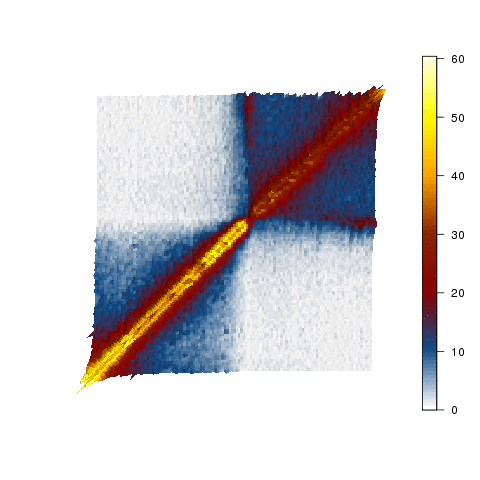
Each cell in the human body must compact two metres of DNA into a nucleus of about ten micrometres, the equivalent of fitting the Eiffel Tower inside a one cent coin. Somehow this is achieved in a way so that the required genes for each cell are accessible to the machinery for their activation. Our team combines genomics approaches (Hi-C and its variants), CRISPR-Cas9 genome editing and microscopy at high temporal resolution to dissect the organization of the genome in space and time, and to see how this is responsible for control of gene expression.
Twitter: @TomSexton_lab
Members
Researchers
PhD students
Engineers
Occasionnal collaborators
Former members
Yousra Ben Zouari (PhD) – went to postdoc at FMI, Basel.
Nezih Karasu (PhD) – went to postdoc at IGMM, Montpellier
Angeliki Platania (PhD)
Sanjay Chahar (postdoc) – went to postdoc at IGMM, Montpellier
Anne Molitor (postdoc) – went to senior engineer position at Institute of Molecular Immuno-Rheumatology, Strasbourg
Natalia Sikorska (Postdoc) – went to medical training
Gui Oliveira (Postdoc) – went to software engineer position with MathWorks
Dominique Kobi (engineer) – went to position with Machery-Nagel
Manon Maroquenne (engineer) – went to position in Osteoarticular Biology, Bioengineering and Bio-imaging lab, Paris
Audrey Mossler (engineer) – went to Imperial College, UK
Current projects
We are interested in all aspects of how 3D or 4D genome organization affects gene expression, but our current research can be split into two major axes:
- Genome-wide molecular biology approaches (e.g. Capture Hi-C) to characterize epigenomic changes accompanying developmental transitions and responses to chemotherapeutic drugs.
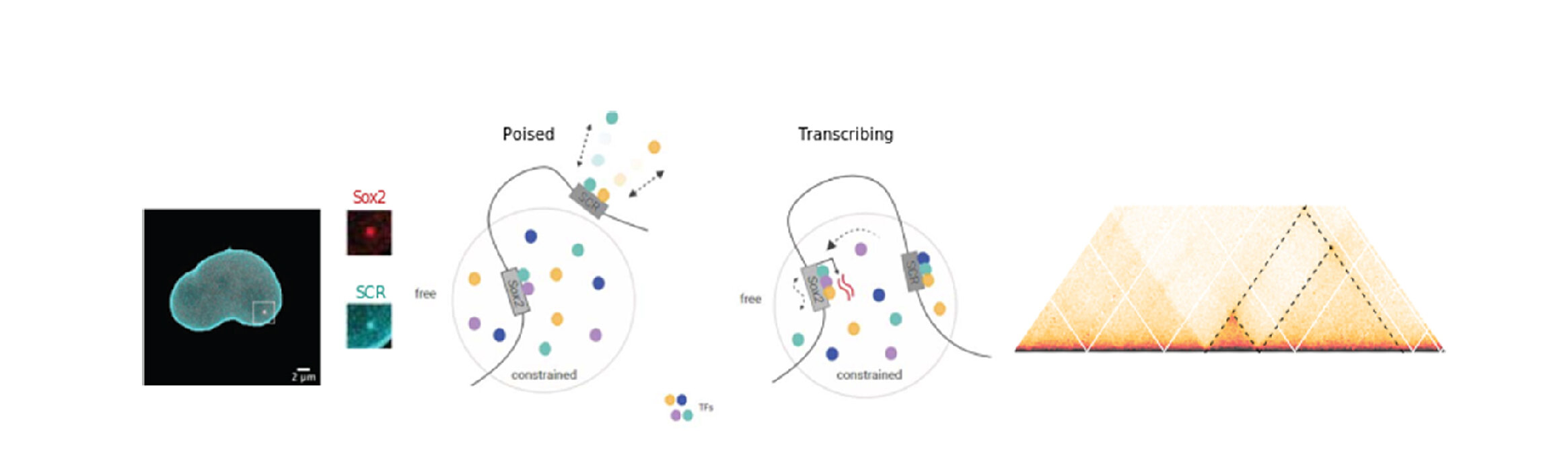
- Tagging specific genomic loci and tracking them in real time to see how chromatin mobility affects (and is affected by) transcription and DNA repair responses.
Our first attempt at modelling the stem cell nucleus.
Collaborations and networks
In IGBMC
- Nacho Molina
- Daniel Metzger
In Strasbourg
- Karine Merienne (Laboratory of Cognitive and Adaptive Neurosciences)
- Jean-Noel Freund/Claire Domon-Dell (Interface of Fundamental and Applied Research in Cancerology)
In France
- Kerstin Bystricky (Centre of Integrative Biology, Toulouse)
- Christophe Ginestier/Emmanuelle Charafe-Jauffret (Cancer Research Centre, Marseille)
- Mounia Lagha (Institute of Molecular Genetics, Montpellier)
- Reini Luco (Institut Curie, Paris)
- Salvatore Spicuglia (Theories and Approaches of Genomic Complexity, Marseille)
- Jean-Christophe Andrau (Institute of Molecular Genetics, Montpellier)
- Daniel Jost (Ecole Normale Superieur, Lyon)
International
- Jennifer Mitchell (University of Toronto, Canada)
- Tineke Lenstra (National Cancer Institute, Netherlands)
- Akis Papantonis (University of Goettingen, Germany)
- Evi Soutoglou (Genome Damage and Stability Centre, UK)
Funding and partners
ERC Starting Grant 2015-2021
ATIP-Avenir 2014-2017
ANR; INCa; USIAS
Awards and recognitions
Claude Paoletti Prize, 2015
Resources
Check out our Github tools :
- For Capture Hi-C analysis and browsing (ChiCMaxima) : github.com/yousra291987/ChiCMaxima
- For browsing 4C data (4See) : github.com/TomSexton00/4See
- For measuring diffusive properties from live imaging results (GPTool): github.com/guilmont/GP-Tool
Publications
2024
Article in a journal
Gene-to-gene coordinated regulation of transcription and alternative splicing by 3D chromatin remodeling upon NF-κB activation
- Paul Marie
- Matéo Bazire
- Julien Ladet
- Lamya Ben Ameur
- Sanjay Chahar
- Nicolas Fontrodona
- Tom Sexton
- Didier Auboeuf
- Cyril F Bourgeois
- Franck Mortreux
Nucleic Acids Research ; Volume: 52 ; Page: 1527-1543
Article in a journal
Transcription processes compete with loop extrusion to homogenize promoter and enhancer dynamics
- Angeliki Platania
- Cathie Erb
- Mariano Barbieri
- Bastien Molcrette
- Erwan Grandgirard
- Marit de Kort
- Wim Pomp
- Karen Meaburn
- Tiegh Taylor
- Virlana Shchuka
- Silvia Kocanova
- Mariia Nazarova
- Guilherme Monteiro Oliveira
- Jennifer Mitchell
- Evi Soutoglou
- Tineke Lenstra
- Nacho Molina
- Argyris Papantonis
- Kerstin Bystricky
- Tom Sexton
Science Advances ; Volume: 10 ; Page: eadq0987
2023
Article in a journal
Transcription induces context-dependent remodeling of chromatin architecture during differentiation
- Sanjay Chahar
- Yousra Ben Zouari
- Hossein Salari
- Dominique Kobi
- Manon Maroquenne
- Cathie Erb
- Anne Molitor
- Audrey Mossler
- Nezih Karasu
- Daniel Jost
- Tom Sexton
PLoS Biology ; Volume: 21 ; Page: e3002424
Article in a journal
Short tandem repeats are important contributors to silencer elements in T cells
- Saadat Hussain
- Nori Sadouni
- Dominic van Essen
- Lan T M Dao
- Quentin Ferré
- Guillaume Charbonnier
- Magali Torres
- Frederic Gallardo
- Charles-Henri Lecellier
- Tom Sexton
- Simona Saccani
- Salvatore Spicuglia
Nucleic Acids Research ; Volume: 51 ; Page: 4845-4866
Pre-publication, Working Document
Competition between transcription and loop extrusion modulates promoter and enhancer dynamics
- Angeliki Platania
- Cathie Erb
- Mariano Barbieri
- Bastien Molcrette
- Erwan Grandgirard
- Marit Ac de Kort
- Karen Meaburn
- Tiegh Taylor
- Virlana M Shchuka
- Silvia Kocanova
- Guilherme Monteiro Oliveira
- Jennifer A Mitchell
- Evi Soutoglou
- Tineke L Lenstra
- Nacho Molina
- Argyris Papantonis
- Kerstin Bystricky
- Tom Sexton
- Guilherme Monteiro Oliveira
2022
Pre-publication, Working Document
Context-dependent transcriptional remodeling of TADs during differentiation
- Sanjay Chahar
- Yousra Ben Zouari
- Hossein Salari
- Anne Molitor
- Dominique Kobi
- Manon Maroquenne
- Cathie Erb
- Audrey Mossler
- Nezih Karasu
- Daniel Jost
- Tom Sexton
Article in a journal
Concurrent CDX2 cis -deregulation and UBTF::ATXN7L3 fusion define a novel high-risk subtype of B-cell ALL
- Marie Passet
- Rathana Kim
- Stéphanie Gachet
- François Sigaux
- Julie Chaumeil
- Ava Galland
- Thomas Sexton
- Samuel Quentin
- Lucie Hernandez
- Lise Larcher
- Hugo Bergugnat
- Tao Ye
- Nezih Karasu
- Aurélie Caye
- Beate Heizmann
- Isabelle Duluc
- Patrice Chevallier
- Philippe Rousselot
- Françoise Huguet
- Thibaut Leguay
- ...
Blood ; Volume: 139 ; Page: 3505-3518
Article in a journal
The histone variant macroH2A1.1 regulates RNA polymerase II-paused genes within defined chromatin interaction landscapes
- Ludmila Recoules
- Alexandre Heurteau
- Flavien Raynal
- Nezih Karasu
- Fatima Moutahir
- Fabienne Bejjani
- Isabelle Jariel-Encontre
- Olivier Cuvier
- Thomas Sexton
- Anne-Claire Lavigne
- Kerstin Bystricky
Journal of Cell Science ; Volume: 135 ; Page: jcs259456
2021
Article in a journal
Age-related and disease locus-specific mechanisms contribute to early remodelling of chromatin structure in Huntington’s disease mice
- Rafael Alcalá-Vida
- Jonathan Seguin
- Caroline Lotz
- Anne Molitor
- Ibai Irastorza-Azcarate
- Ali Awada
- Nezih Karasu
- Aurélie Bombardier
- Brigitte Cosquer
- Jose Luis Gomez Skarmeta
- Jean-Christophe Cassel
- Anne-Laurence Boutillier
- Thomas Sexton
- Karine Merienne
Nature Communications ; Volume: 12 ; Page: 364
Article in a journal
Precise measurements of chromatin diffusion dynamics by modeling using Gaussian processes
- Guilherme Oliveira
- Attila Oravecz
- Dominique Kobi
- Manon Maroquenne
- Kerstin Bystricky
- Thomas Sexton
- Nacho Molina
Nature Communications ; Volume: 12 ; Page: 6184
