Molecular Basis for Protein Synthesis by the Ribosome
Team Leader : Gulnara YUSUPOVA
Department : Integrated structural biology
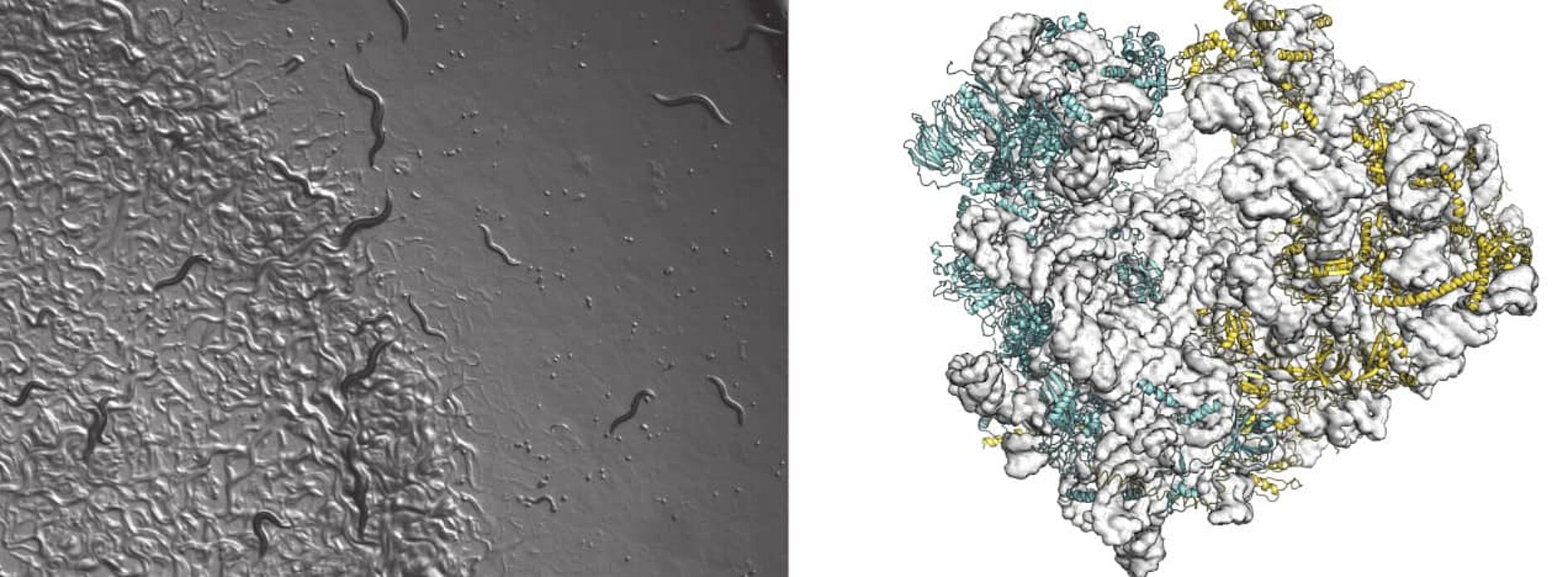
Recently, ribosome heterogeneity has emerged as a valid idea with many scientific groups presenting evidence corroborating compositional diversity of the ribosome. These differences can arise in many ways: ribosomal protein content, preferential utilization of paralogues, sequence variation of ribosomal RNA, post-translational modifications of RP and rRNA have all been documented. Although the effects of ribosome heterogeneity still mostly remain a mystery, emerging studies indicate that the variations in ribosome composition may underlie several vital biological functions such as translational control or environmental responses.
We study how cells exploit this ribosome heterogeneity to regulate translation during development in complex multicellular organisms and how ribosome composition can differ depending on the developmental stage of the cell. To answer these questions, we use an innovative combination of biochemistry and cryo Electron Microscopy for structural characterization of ribosomal complexes taken at different developmental stages and study their phenotypic expression and function in the nematode Caernorhabditis elegans.
80S ribosomes are isolated from C. elegans at different stages of development. An isolation protocol was established for obtaining 80S C. elegans ribosomes suitable for structural investigations, and recently a high resolution cryo-EM structure has been determined (2.6Å resolution GSFSC=0.134) of the L3 developmental stage. More structures will be determined of ribosomes from the different developmental stages and careful comparison will be performed to investigate ribosome heterogeneity, which will then subsequently be exploited by our collaborative partner Dr. Sophie Jarriault to dissect through genetic and functional in vivo studies how changes in ribosome content affect cellular homeostasis, identity and ultimately animal development.
Sophie Jarriault. IGBMC
Team Leader : Gulnara YUSUPOVA
Department : Integrated structural biology