Regulation of transcription
Regulation of transcription
The blueprint of all cellular organisms is stored in the form of genes in their DNA. This genetic information needs to be accessed, retrieved, and decoded in a process called gene expression. Gene expression needs to occur in a precisely controlled manner in order to convert the genotype of an organism to the phenotype of an organism. Erroneous gene expression on the other hand has severe consequences. The goal in our team is to obtain a detailed and mechanistic understanding of the cellular machineries, which carry out gene expression. This is important for several reasons:
1) Gene expression plays a fundamental role and affects almost every aspect of biology. A lot of our motivation to understand gene expression is therefore driven by curiosity.
2) The majority of drugs against human pathogens target the enzymes involved in gene expression. The better you understand your target, the better you understand how to improve drugs or design new ones.
3) Many human pathologies including cancers, neurodevelopmental disorders or autoimmune diseases are the result of misregulated gene expression. A prerequisite to developing effective strategies against a specific disease, however, is to understand the underlying process in as much detail as possible.
The first step of gene expression is to transcribe DNA into RNA and it is carried out by a universally conserved enzyme called RNA polymerase. The DNA transcript, called messenger RNA or mRNA, is subsequently translated to protein by another universally conserved molecular machine called the ribosome. Many organisms including humans need to process the mRNA first before it can be translated.
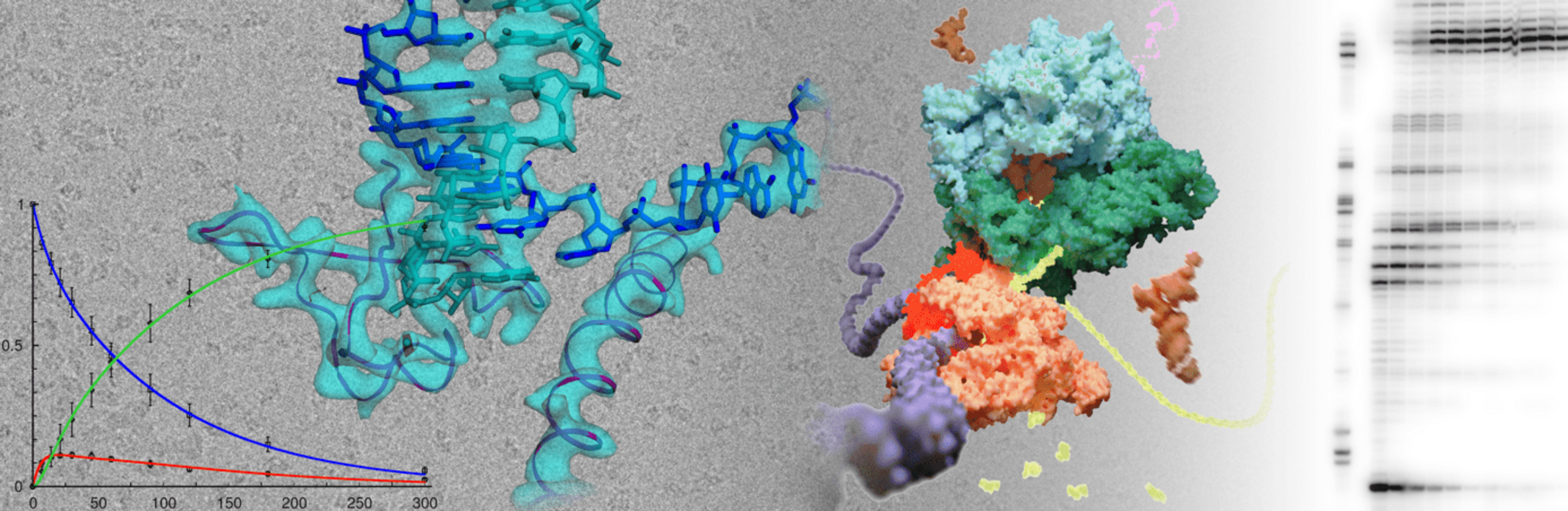
Gene expression is tightly controlled at each step but transcription is arguably one of the most important targets for regulatory processes. One focus of research in our team is to understand how proteins called transcription factors are able to regulate RNA polymerase and therefore modulate the transcription of DNA to RNA. Transcription is often directly regulated by RNA and we would like to understand how regulatory RNA elements affect RNA polymerase. Finally, the enzymes carrying out gene expression do not operate in isolation. They are often organized in larger, supramolecular assemblies. In these assemblies their functions are coordinated and new functions can emerge. We are increasingly addressing this aspect and study how RNA polymerase is organized along other key enzymes in higher order structures.
We use bacterial or eukaryotic models and combine molecular biology, and biochemistry with X-ray crystallography and single particle Cryo-EM to describe these processes at the molecular level.
Members
Researchers
Post-doctoral fellows
PhD students
Engineers
Former members
- Jinal Shukla, 2014-2016
- Xieyang Guo, 2014-2018
- Mo’men Abdelkareem, 2015-2020
- Chenghin Zhu, 2015-2021
- Claire Batisse, 2016-2020
- Dmitro Rodnin, 2016-2017
- Flore Kersten, 2017
- Ayesha Dinshaw Eduljee, 2018-2021
Current projects
- Structural investigations on transcription factors bound to RNA polymerase elongation complexes in states of transcriptional pausing
- Non-coding RNAs modulating RNA polymerase elongation complexes
- Structural basis for the coupling of transcription and translation
- Interplay between DNA topology and transcription
- Coupling of transcription and mRNA processing (Project leader Clément CHARENTON)
Collaborations and networks
- We work with Terence STRICK (Ecole Normale Supérieure de Paris) to combine structural studies with single molecule techniques to elucidate fundamental processes in transcription regulation
- We work with Olivier ESPÉLIE (College de France) and Valérie LAMOUR (IGBMC) to study the interplay between transcription and DNA topology
- We work in a collaborative atmosphere with different teams at IGBMC
News

From DNA to Proteins: A Perfectly Orchestrated Molecular Dance
Gene expression involves the transcription of DNA into mRNA by RNA polymerase and the translation of this mRNA into proteins within the ribosome. In…
Read more
Funding and partners
 ERC
ERC ANR
ANR  Fondation Bettencourt-Schueller
Fondation Bettencourt-Schueller Ligue contre le Cancer
Ligue contre le Cancer Programme d'investissement avenir
Programme d'investissement avenir
Awards and recognitions
- ATIP-Avenir 2014
- ERC Starting Grant 2015
- Prix Guy Ourisson 2018
- Coups d’élan pour la recherche française 2021
Publications
2025
Article in a journal
DNA topoisomerase I acts as supercoiling sensor for bacterial transcription elongation
- Vita Vidmar
- Céline Borde
- Lisa Bruno
- Nataliya Miropolskaya
- Maria Takacs
- Claire Batisse
- Charlotte Saint-André
- Chengjin Zhu
- Olivier Espéli
- Valérie Lamour
- Albert Weixlbaumer
Nature Structural and Molecular Biology
2024
Article in a journal
DNA topoisomerase I acts as supercoiling sensor for transcription elongation in E. coli
- Vita Vidmar
- Céline Borde
- Lisa Bruno
- Maria Takacs
- Claire Batisse
- Charlotte Saint-André
- Chengjin Zhu
- Olivier Espéli
- Valérie Lamour
- Albert Weixlbaumer
BioRxiv
2022
Article in a journal
Structural insights into RNA-mediated transcription regulation in bacteria
- Sanjay Dey
- Claire Batisse
- Jinal Shukla
- Michael Webster
- Maria Takacs
- Charlotte Saint-André
- Albert Weixlbaumer
Molecular Cell ; Volume: 82 ; Page: 3885-3990
2019
Article in a journal
Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation
- Mo'Men Abdelkareem
- Charlotte Saint-André
- Maria Takacs
- Gabor Papai
- Corinne Crucifix
- Xieyang Guo
- Julio Ortiz
- Albert Weixlbaumer
Molecular Cell ; Volume: 75 ; Page: 298 - 309.e4
2018
Article in a journal
Structural Basis for NusA Stabilized Transcriptional Pausing
- Xieyang Guo
- Alexander G. Myasnikov
- James Chen
- Corinne Crucifix
- Gabor Papai
- Mária Takács
- Patrick Schultz
- Albert Weixlbaumer
Molecular Cell ; Volume: 69 ; Page: 816-827.e4

