mRNA processing
mRNA processing
The blueprint to synthetize the building blocks of living organisms, the proteins, is contained in their DNA. Nevertheless, this information is not directly decoded from DNA by the protein-synthesis machinery but first copied as transient and mobile copies of the genetic material: the messenger RNAs (mRNA).
The use of an intermediate molecule not only protects the precious hereditary information safeguarded in DNA but also offers plasticity to fine tune gene expression.
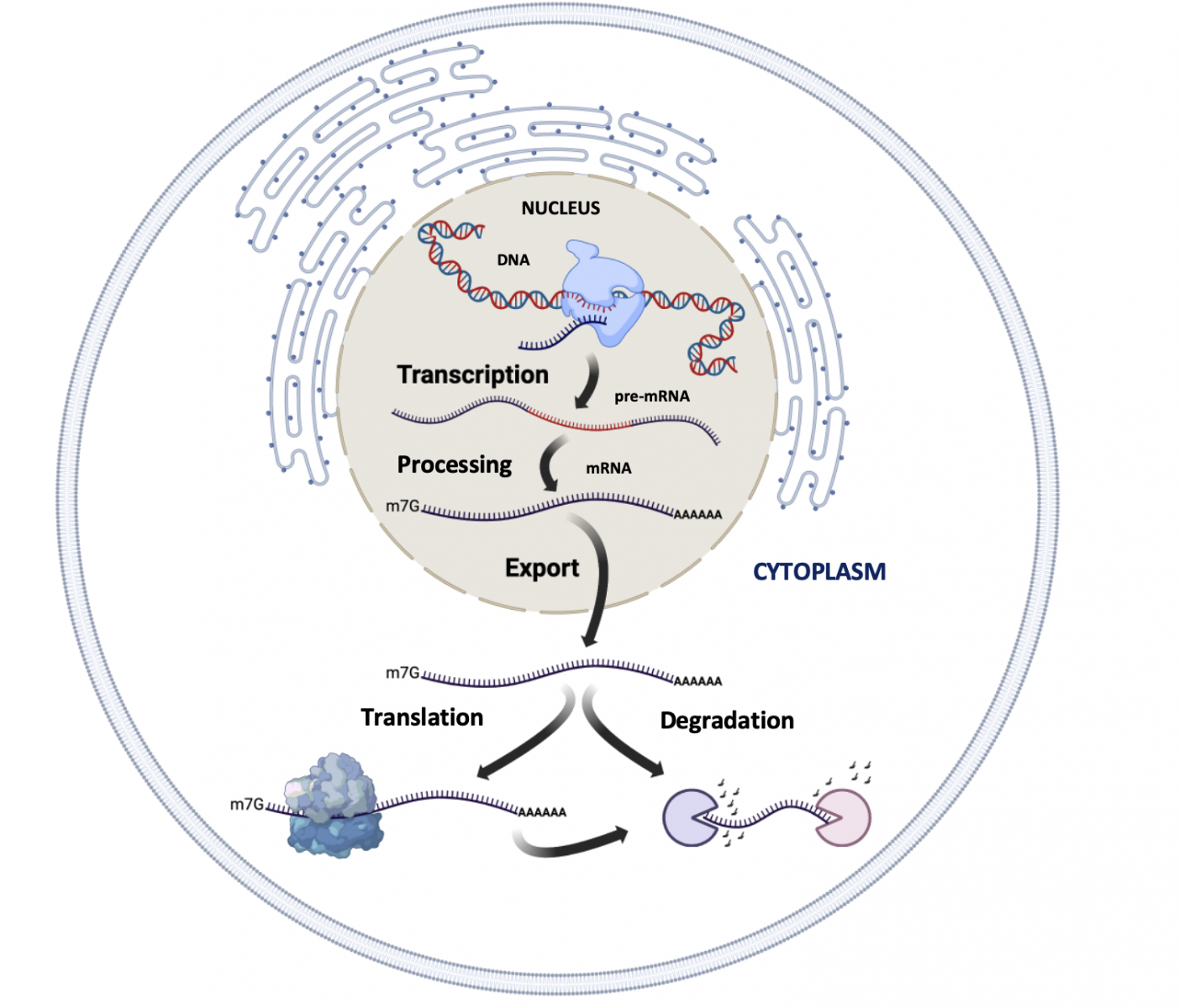
The life cycle of eukaryotic mRNAs is complex. First, they are synthetised during transcription by the RNA polymerase II (Pol II). Second, they are modified co/post-transcriptionally during the mRNA processing step. Third, the resulting mature mRNAs are transported in the cytoplasm, via nuclear export mechanisms, to be decoded into proteins by the ribosome during translation. Finally, the transcripts are degraded by specific mRNA decay pathways.
The team is interested in the mRNA processing step. In particular, we aim to determine the mechanisms underlying the function of molecular machines that modify mRNAs, changing their stability, their "translatability" or even the genetic information they contain!
For this, the molecular complexes of interest, composed of proteins and/or RNA are reconstituted from recombinant sources and/or cell extract. Their chemical compositions and structures are then studied using biochemical, biophysical and structural biology approaches (electron cryo-microscopy (cryo-EM), in particular).
Current projects
Splicing fidelity
During splicing, the spliceosome identifies and then eliminates particular sequences present within precursor messenger RNAs (pre-mRNA), the introns. For a given pre-mRNA, the sequences removed may vary depending on cell type or various signals. This "alternative splicing" phenomenon enblaes, from a single gene, to produce several types of proteins!
Splicing must be performed without error to avoid the production of aberrant mRNAs encoding potentially toxic proteins. Splicing fidelity relies on the precision of the spliceosome, a large molecular machinery composed of dozens of proteins and RNAs, which defines the boundaries of introns before catalyzing their excisions.
This project aims to determine how the spliceosome selects introns without making errors, while showing great flexibility to allow splicing changes during alternative splicing. For this, the human spliceosome will be isolated at key moments when it recognizes its target sequences in order to study its composition and three-dimensional structure.
Funding and partners
ERC Starting Grant

ATIP-avenir

News

A new research team specialized in RNA maturation
Clément Charenton, who was temporarily welcomed in Albert Weixlbaumer's team, is starting his own research team at the IGBMC in the Department of…
Read more
