Activated transcription
Subgroup Leaders : Adam BEN SHEM , Gabor PAPAI
Teams : Transcription co-activators
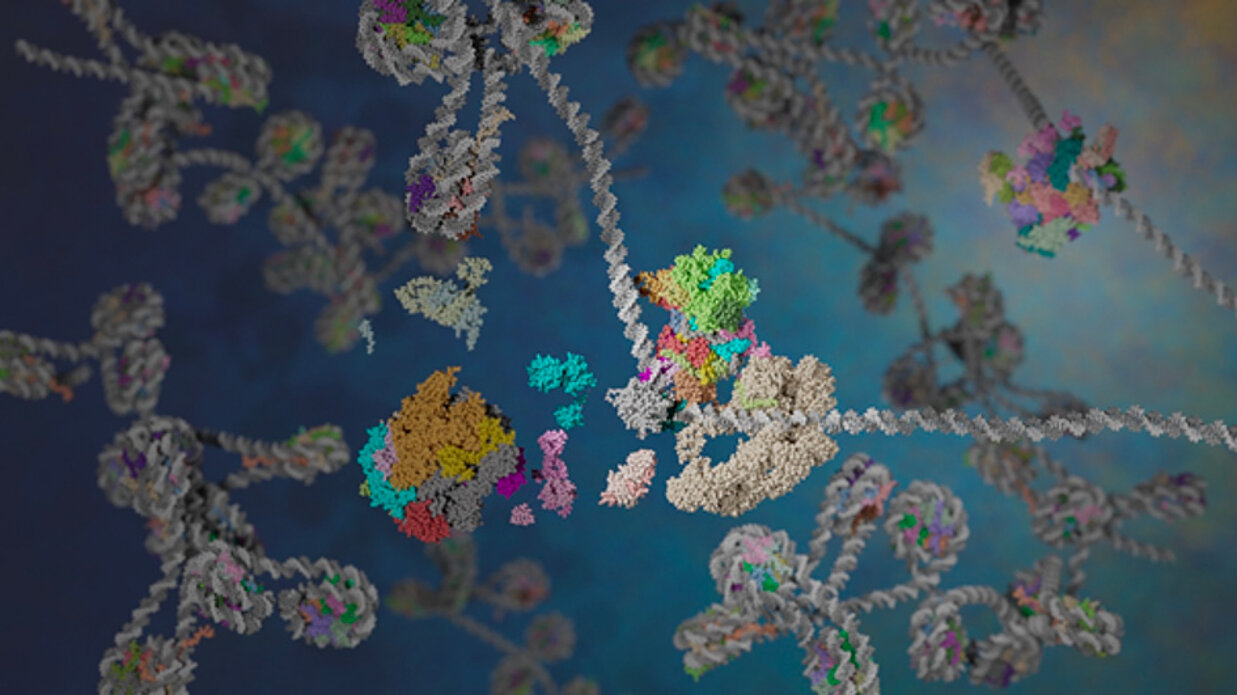
Our group aims at understanding the mechanisms of gene expression at the transcriptional level and to decipher the various degrees of organization that determine the fine tuning of transcription at the initiation stage.
In Eukaryotes transcription of protein encoding genes is initiated by the controlled deposition of the TATA-box binding protein TBP onto gene promoters, followed by the ordered assembly of a preinitiation complex composed of seven General Transcription Factors (GTF) together with RNA polymerase II (Pol II). In vivo this process occurs in the context of chromatin in which DNA is organized into nucleosomes, a general repressive state where regulatory sequences are partially occluded. Furthermore, sequence-dependent activator proteins bind upstream of the core promoter in order to adapt mRNA synthesis levels to stress or environmental cues. Activator proteins recruit multi-functional co-activator complexes which modify the structure of chromatin, act as bridging or stabilizing factors with GTFs and help depositing TBP on gene promoters. Co-activator action is not only required for transcription initiation, but these complexes also play a major role in the different DNA repair pathways.
We investigate the structure and function of the yeast transcriptional coactivator complexes TFIID and SAGA, which bind TBP and load it onto gene promoters in a highly controlled way. These two complexes contain a similar TBP-binding machinery composed of a deformed octamer of histone-fold containing proteins. The second class a co-activators studied in our team is modifying the structure of chromatin by acetylating histone tails and other nuclear proteins. SAGA and NuA4 are prototypical examples of coactivators containing a Histone Acetyl Transferase activity coupled to other functions, specific to each complex. More recently our team addressed the structural organization of a third class of coactivators which remodels nucleosomes in an ATP-dependent way. We are studying the SWI/SNF chromatin remodeler family in order to understand their mode of action and how they are recruited to their genomic target. The function of these molecular assemblies is coupled to conformational changes and to allosteric regulation mechanisms. We aim at analyzing the dynamic behavior of co-activators by combining high resolution cryo-Electron Microscopy and FRET experiments.
Transcription and DNA repair are two fundamental biological processes which have a high impact on human health, in particular for our understanding of tumor origin and progression. Apparently distinct, these two processes are indeed interconnected and share the same molecular mechanisms. The transcription/repair factor TFIIH has long been identified as a common actor bridging the initiation step of transcription and the nucleotide excision repair (NER) pathway by providing the helicases required to unwind the DNA around lesions in order to have them repaired. More recently this interconnection between the transcription and the repair machinery appeared even more intricate since all the repair factors were found to be recruited during important transcriptional switches in cell differentiation or reprogramming. Although recent breakthrough structural and biophysical studies have led to a marked progress regarding TFIIH architecture, our understanding of its different functional states and interactions with partners in transcription and DNA repair are still limited.
Vertebrate coactivators play important roles in development, cell differentiation, STEM cell maintenance, apoptosis or aging, and are involved in several genetic diseases and cancer. To unravel the mechanisms of gene expression and their deregulation in human pathologies, we develop purification strategies to study the human complexes. High resolution structures of human co-activators are required to shed light on their molecular mechanisms, help understand the role of mutations found in diseases and deliver a framework to evaluate the action of inhibitors of enzymatic activities or protein-protein interactions.
We are using a multi scale structural biology approach to study Co-activators which are fully active in a chromatin environment. We reconstitute in vitro nucleoprotein systems assembled on chromatin templates and determine their structure by single particle cryo-Electron Microscopy. We also aim at analyzing the localization, the structure and the molecular environment of transcription factors in the context of a fully functional cell nucleus. We analyze the dynamic behavior of these complexes in living cells by fluorescence microscopy approaches. We probe the nuclear environment of these complexes by cryo-electron tomography of cryo immobilized cells and by using antibody-coupled golds probes to locate the proteins of interest.
To address these challenging questions, we implement and benefit of the most advanced technology developments and participate actively in new methodological advances in specimen preparation (expression systems in mammalian cells, cryo sectioning methods, high pressure freezing protocols), cryo-electron microscopy (new detectors, ultra-stable cold stages, automated data collection, Cs correctors, energy filtered images, phase plates, FIB/SEM imaging), image analysis (parallelized computer grids, new algorithms for 3-D alignments or segmentation, sorting of heterogeneous datasets) and data integration (correlative high resolution light and electron microscopy, combination of heterologous information).
Subgroup Leaders : Adam BEN SHEM , Gabor PAPAI
Teams : Transcription co-activators
Subgroup Leader : Arnaud POTERSZMAN
Teams : Transcription co-activators
The assembly of the transcription preinitiation complex (PIC), composed of general factors (GTFs) and the Pol II enzyme, on gene promoters is a key regulatory step that selects the gene to be expressed. PIC assembly on a TATA-box containing promoter has been reconstituted in vitro and involves the interaction of the TATA Binding Protein (TBP) with its cognate promoter sequence, the subsequent binding of the GTFs, TFIIA and TFIIB, that recruit Pol II with the remaining GTFs: TFIIE, TFIIF and TFIIH. Close to atomic resolution structures of this in vitro reconstituted PIC have been determined recently.
However, in vivo TBP associates with 13 (14 in yeast) TBP-associated factors (TAFs) and forms the transcription factor IID (TFIID) that not only nucleates PIC formation, but has been suggested to function also as a transcriptional co-activator by interacting with transcription activators. Importantly, the structure of TFIID-containing PICs is not known, leaving open the possibility that the composition and architecture of in vivo assembled PICs may differ from that reconstituted in vitro.
To better understand the role of native TFIID in endogenous PIC formation and consequent transcription initiation we aim at:
This information combined with genome-wide TAF, TBP, GTF and Pol II occupancy and gene expression profiling data sets will identify distinct sets of TFIID-containing PIC assemblies on different promoter classes that are involved in transcription initiation.
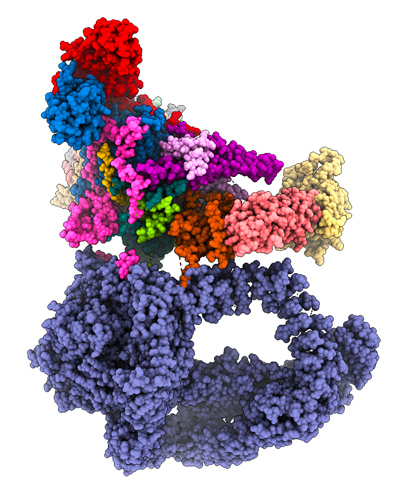
Our goal is to understand the functional role, the structural organization and the mode of action of the transcriptional co-activator SAGA; a multi protein complex containing 19 different subunits with for a total molecular weight of 1.9 MDa. SAGA contains two enzymatic activities to acetylate or deubiquinate nucleosomal histone tails by the GCN5 and USP22 subunits, respectively. Impairment of either of SAGA’s enzymes, GCN5 or USP22, leads to embryonic lethality and mutations have been described in human genes encoding these subunits that cause neurodegeneration or contribute to tumorigenesis. A dedicated module of SAGA is responsible for its interaction with the TATA-box binding protein (TBP), a key component of the PIC. The large 400 kDa Tra1 subunits is involved in transcriptional activator binding thus making the link with the cellular signaling pathways.
Our team played key roles in the structural characterization of human and yeast SAGA. However, the impact of each individual SAGA activity on gene expression is presently poorly understood. We aim at understanding the effects of chromatin recognition, catalytic activities, activator recognition and TBP binding on the efficiency of gene transcription. The mechanism by which activator binding to Tra1 triggers PIC assembly and increases mRNA production is currently unknown, and the activator-dependent recruitment of SAGA to chromatin likely involves coordinated conformational changes yet to be characterized. An important question in the field is to understand how SAGA can regulate transcription initiation both positively and negatively. A striking example of this dual role of SAGA is found in the fission yeast S. pombe, where cells stop proliferation in response to nutrient starvation and differentiate into quiescent spores. Distinct SAGA subunits play opposing roles at the promoters of differentiation genes, depending on nutrient availability. In rich conditions, SAGA represses these promoters, whereas upon nutrient starvation, it activates the transcription of the same genes. We aim at uncovering the molecular mechanism by which SAGA integrates environmental signals to regulate gene expression either positively or negatively.
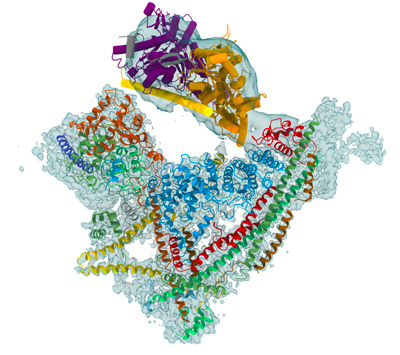
Chromatin architecture is a key level of organization that regulates genome expression, DNA replication, maintenance, and epigenetic control. Chromatin remodelers use the energy of ATP hydrolysis to act on the fundamental unit of chromatin, the nucleosome, and destabilize the interaction of the histone octamer with DNA. The action of remodelers allows nucleosomes to be phased with respect to their neighbors, to be repositioned, exchanged or evicted from key regulatory DNA sequences. Remodelers regulate the access of specific molecular machines to genomic DNA sequences and thereby modulate the main processes in the nuclei including transcription, replication and DNA repair.
The yeast SWI/SNF remodeler complex has a molecular weight of about 1.1 MDa. It comprises the catalytic Snf2 subunit, a helicase which couples the energy derived from ATP hydrolysis into chromatin remodeling, as well as 10 additional subunits. While the function of the Snf2 helicase is well documented, little is known about the role of the additional subunits. Despite the key biological importance of chromatin remodelers, we are currently missing a structural framework to understand the ATP-dependent mechanism by which the SWI/SNF complex mobilizes or disrupts the nucleosome. Several models have been proposed for the nucleosome sliding mechanism and the eviction reaction is still under debate.
The removal of promoter nucleosomes to form a nucleosome depleted region that favors transcription initiation is a generally accepted mechanism to explain the positive effect of SWI/SNF on gene expression. An important open question is to understand how SWI/SNF is directed to its functional sites in the genome. The Swi2, Swi5 and Swi1 subunits were shown to interact with transcriptional activators and may recruit the remodeler to gene promoters in response to cellular signaling pathways. Alternatively the remodeler can also be recruited by taking advantage of epigenetic marks on histones tails. In particular, the Snf2 bromodomain is responsible of the activator independent anchoring of SWI/SNF to acetylated promoter nucleosomes. The molecular details of these interactions are poorly described and the physical coupling between the recruitment and remodeling activities is presently not understood.
A core of SWI/SNF subunits is conserved during evolution, but the complex has diversified from yeast to mammals to reach a molecular mass of 2 MDa in the human PBAF and BAF complexes. The subunit composition is cell type specific due to the differential expression of paralogous genes for several subunits. It is generally accepted that different subunit combinations confer cell specificity and allow SWI/SNF to interact with tissue-specific transcription factors to regulate the expression of target genes. High-resolution structural data of endogenous human BAF complexes would be a major advance in understanding the mechanism of action of the human complexes, and their role in chromatin targeting.
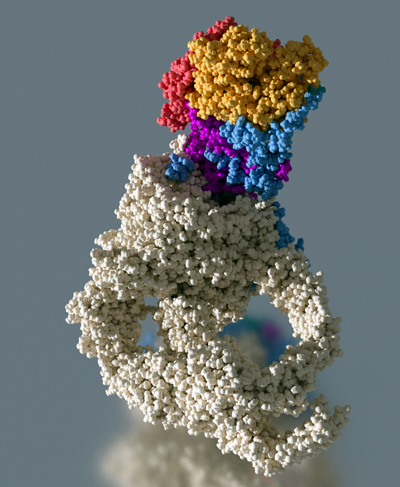
Our goal is to understand the functional role, the structural organization and the mode of action of the yeast transcriptional co-activator NuA4, a multi protein complex containing 13 different subunits with for a total molecular weight of 1.0 MDa. NuA4 integrates epigenetic signaling by reading and writing post-translational histone modifications, interacts with transcriptional activators and incorporates specific histone variants. The Esa1 subunit of NuA4 is an acetyl transferase capable of modifying histone H4 and is essential for yeast growth. The large 430 kDa Tra1 subunit is involved in transcriptional activator binding thus making the link with the cellular signaling pathways. NuA4 also participates in the recruitment of the histone variant H2A.Z, a chromatin marker that bookmarks active gene promoters. To perform this function NuA4 interacts with the chromatin remodeler SWR1. In humans, the homologous NuA4 and SWR1 are physically associated to form the TIP60/p400 complex, which encompasses both histone acetyltransferase (TIP60) and histone exchange (p400) activities.
The structural organization of the full NuA4 and SWR1 complexes is poorly understood and we aim at solving their high-resolution structure by cryo-EM. The mechanism by which these factors are recruited to gene promoters is currently unknown and we study supramolecular complexes between NuA4 and, both transcriptional activators and nucleosomes to understand the network of interactions that target NuA4 to active genes. The histone H2A.Z exchange activity has not been studied so far at the structural level. We study the structure of the NuA4 and SWR1 complexes by cryo-EM as well as structural intermediates of the exchange reaction. We analyze the structure and the function of the homologous human TIP60/p400 complexes in order to better understand the H2A.Z exchange mechanism.
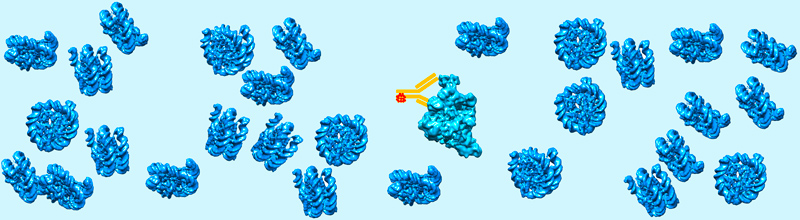
The functions of biological macromolecules are generally studied after their purification and isolation from the other cellular components. This approach is reductionist since important properties such as membrane-less compartmentalization, phase separation, condensate formation, weak or transient interactions are only occurring in the crowded environment of the intact cell. Concepts familiar to polymer scientists drive or regulate important cellular processes such as gene expression or cell division by creating dynamic microenvironments with dedicated functional properties. These phenomena have been described by life cell imaging but need to be characterized at the molecular level. Cryo-EM approaches have been developed to visualize molecular assemblies in their natural cellular environment. Optimal “close-to-native” specimen preparation conditions can be achieved using cryo-immobilization techniques and tri-dimensional (3-D) reconstruction of the cell can be performed by cryo-Electron Tomography (cryo-ET) of thin cellular slices produced by cryo-sectioning or focused ion beam (FIB) milling of lamellae. The possibility to visualize molecular complexes in their native cellular environment opens new avenues of research to understand their cellular function.
A current challenge is to identify macromolecular assemblies of low cellular abundance or too small to be recognized by their shape. To address this challenge we synthesized in collaboration with Dr Guy Zuber (UMR7242), novel gold-based probes to specifically identify the molecules of interest. The gold particle are coupled to antibody derivatives, delivered into living cells in order to selectively label the target proteins. The electron-dense gold particles will be detected in cell sections by cryo-electron tomography.

Functionalized gold labels to visualize biomolecules and ligands by electron microscopy
A collaborative project to develop new electron dense probes to detect and identify biomolecules in their cellular environment.
In collaboration with Dr Guy ZUBER (UMR7242, ESBS, Illkirch, france) and Dr Ovidiu ERSEN (UMR7504, IPCMS, Strasbourg, France)

This French national multi-disciplinary workforce aims at a providing a multi-scale characterization of the oncogenic cascade, from the atomic level to the dynamic organization of the cell in response to genetic mutations, environmental changes or epigenetic modifications. fr.nanotumor.fr
Aviesan Federative Program (PFA) - Towards the Tumor Cell Atlas
A research program coordinated by Jacky G. Goetz (U1109 - Strasbourg) and Patrick Schultz (IGBMC - Illkirch), and composed of 13 teams working on four axes:
Axis 1: Molecular and structural description of multiprotein complexes initiating the oncogenic process
Axis 2: Establishment of a subcellular morphological atlas during carcinogenesis: From organelles to molecular networks that drive cancer
Axis 3: Biosensors, protein and chemical inhibitors
Axis 4: In vivo integration of targetable candidate protein complexes in patient samples
Ligue contre le cancer labelled team
Structure of multi-protein complexes involved in transcription, epigenetic regulation and DNA repair.

CNRS program 80 Prime
Gold labels to visualize protein complexes in cells by cryo-electron tomography

Aviesan for the Nanotumor project
![]()
![]()
USIAS Fellowship / Structure of human transcription co-activators

PICen project : Structure and function of endogenous TFIID-containing complexes
![]()
Grandes avancées françaises en biologie 2021

Instruct –ERIC : European research infrastructure for structural biology
![]()
Scientific Reports ; Volume: 12 ; Page: 2030
iScience ; Volume: 25 ; Page: 105357
iScience ; Volume: 25 ; Page: 105357
Insoluble Proteins ; Volume: 2406 ; Page: 281-317
Scientific Reports ; Volume: 12 ; Page: 2030
Multiprotein Complexes ; Volume: 2247 ; Page: 39-57
Multiprotein complexes ; Page: 17-38
Nanoscale Advances ; Volume: 3 ; Page: 6940-6948
Journal of Molecular Biology ; Volume: 433 ; Page: 166964
FEBS Journal ; Volume: 288 ; Page: 3135-3147