Large complexes involved in gene expression
Large complexes involved in gene expression
Transcription and translation are fundamental molecular mechanisms of gene activity regulation with profound implications for human health. The ligand-dependent transcriptional regulation by nuclear receptors bound to DNA response elements involves the transient assembly of large co-regulator complexes. These trigger chromatin remodeling and facilitate the assembly of the general transcription machinery on the promoter of the target gene. Gene expression is also regulated at the level of protein synthesis, for example, by protein factors that bind to the ribosome during the translation initiation, elongation and termination phase. The initiation phase is strongly regulated by factors and also by the mRNA itself and well-characterized reaction intermediates of the initiating ribosomal nano-machinery are potential targets for antibiotics. Both transcription and translation complexes represent large, transient macromolecular assemblies that we investigate by using an integrative structural biology approach with crystallography and cryo electron microscopy forming the core.
Current projects
Team members
RESEARCHERS
- Bruno KLAHOLZ
- Ottilie von LOEFFELHOLZ
- Alexandre OURJOUMTSEV
TECHNICIANS
- Isabelle HAZEMANN
POST-DOCTORAL FELLOWS
- Antony LECHNER
- Nadine WERNER
- Samuel HOLVEC
- Roberto BAHENA CERON
- Charles BARCHET
- Sylvain GUGGENBUHL
- Diego LEONARDO
PhD STUDENTS
- Yung-Chieh CHEN
- Sylwan BONY
- Rakhshana BALA KRISHNAN
MASTER STUDENTS
- Cristina PROTUC
Publications
D. C. Mendonça, S. T. B. Morais, H. Ciol, A. P. A. Pinto, D. A. Leonardo, H. D'Muniz Pereira, N. F. Valadares, R. V. Portugal, B. P. Klaholz, R. C. Garratt, A. P. U. Araujo. Structural Insights into Ciona intestinalis Septins: complexes suggest a mechanism for nucleotide-dependent interfacial cross-talk. J Mol Biol2024, 436, 168693. https://doi.org/10.1016/j.jmb.2024.168693
S. Holvec, C. Barchet, A. Lechner, L. Fréchin, S. N. T. De Silva, I. Hazemann, P. Wolff, O. von Loeffelholz & B. P. Klaholz. The structure of the human 80S ribosome at 1.9 Å resolution reveals the molecular role of chemical modifications and ions in RNA. Nat Struct Mol Biol., 2024. https://doi.org/10.1101/2023.11.23.568509 & https://doi.org/10.1038/s41594-024-01274-x See also Research Briefing: https://www.nature.com/articles/s41594-024-01275-w
L. Urzhumtseva, C. Barchet, B. P. Klaholz & A. G. Urzhumtsev. Program VUE: analysing distributions of cryo-EM projections using uniform spherical grids. J. Appl. Cryst., 2024, 57. https://doi.org/10.1107/S1600576724002383
Y. Liao, L. Andronov, X. Liu, J. Lin, L. Guerber, L. Lu, A. Agote-Arán, E. Pangou, L. Ran, C. Kleiss, M. Qu, S. Schmucker, L. Cirillo, Z. Zhang, D. Riveline, M. Gotta, B. P. Klaholz, I. Sumara. UBAP2L ensures homeostasis of nuclear pore complexes at the intact nuclear envelope. J Cell Biol, 2024, 223, e202310006. https://doi.org/10.1083/jcb.202310006
R. Bahena-Ceron, C. Teixeira, J. R. J. Ponce, P. Wolff, F. Couzon, P. François, B. P. Klaholz, F. Vandenesch, P. Romby, K. Moreau and S. Marzi. RlmQ: A Newly Discovered rRNA Modification Enzyme Bridging RNA Modification and Virulence Traits in Staphylococcus aureus. RNA, 2024, 30, 200-212. https://doi.org/10.1261/rna.079850.123
P.V. Afonine, P.D. Adams, O.V. Sobolev, A.G. Urzhumtsev. Accounting for nonuniformity of bulk-solvent: A mosaic model. Protein Sci.2024, 33, e4909. https://doi.org/10.1002/pro.4909
A. Urzhumtsev, P. Adams, P. Afonine. Universal parameters of bulk-solvent masks. Acta Crystallogr A Found Adv.2024, 80, 194-201. https://doi.org/10.1107/S2053273324000299
C. Barchet, L. Fréchin, S. Holvec, I. Hazemann, O. von Loeffelholz & B. P. Klaholz. Focused classifications and refinements in high-resolution single particle cryo-EM analysis. J Struct Biol., 2023, 215, 4, 108015. https://doi.org/10.1016/j.jsb.2023.108015
A. Cappannini, A. Ray, E. Purta, S. Mukherjee, P. Boccaletto, S. N. Moafinejad, A. Lechner, C. Barchet, B. P. Klaholz, F. Stefaniak and J. M. Bujnicki. MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res., 2023, 1-6. https://doi.org/10.1093/nar/gkad1083
A. K. Mohideen Patel, P. Vilela, T. Baba Shaik, A. G. McEwen, I. Hazemann, K. Brillet, E. Ennifar, A. Hamiche, G. V. Markov, V. Laudet, D. Moras, B. P. Klaholz and I. M. L. Billas. Asymmetric Dimerization in a Transcription Factor Superfamily is Promoted by Allosteric Interactions with DNA. Nucleic Acids Res., 2023, 51, 8864-8879. https://doi.org/10.1093/nar/gkad632
S. Postel, L. Wissler, C. A. Johansson, A. Gunnarsson, E. Gordon, B. Collins, M. Castaldo, C. Köhler, D. Öling, P. Johansson, L. Fröderberg Roth, B. Beinsteiner, I. Dainty, S. Delaney, B. P. Klaholz, I. M. L. Billas & K. Edman. Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication. Nat Struct Mol Biol., 2023, 30, 286-295. https://doi.org/10.1038/s41594-022-00914-4
R. Vuillemot, A. Mirzaei, M. Harastani, I. Hamitouche, L. Fréchin, B. P. Klaholz, O. Miyashita, F. Tama, I. Rouiller, S. Jonic. MDSPACE: Extracting continuous conformational landscapes from cryo-EM single particle datasets using 3D-to-2D flexible fitting based on Molecular Dynamics simulation. J Mol Biol, 2023, 1, 435, 167951. https://doi.org/10.1016/j.jmb.2023.167951
J. A. Louro, R. Boopathi, B. Beinsteiner, A. K. Mohideen Patel, T. C. Cheng, D. Angelov, A. Hamiche, J. Bednar, S. Kale, B. P. Klaholz, S. Dimitrov. Nucleosome dyad determines the H1 C-terminus collapse on distinct DNA arms. Structure, 2023, 31, 1-12. https://doi.org/10.1016/j.str.2022.12.005
L. Fréchin, S. Holvec, O. von Loeffelholz, I. Hazemann & B. P. Klaholz. High-resolution cryo-EM performance comparison of two latest-generation cryo electron microscopes on the human ribosome. J Struct Biol., 2023, 215, 1, 107905. https://doi.org/10.1016/j.jsb.2022.107905
L. Andronov, R. Genthial, D. Hentsch, B. P. Klaholz. splitSMLM, a spectral demixing method for high-precision multi-color localization microscopy applied to nuclear pore complexes. Commun Biol, 2022,1-13. bioRxiv:https://doi.org/10.1101/2021.12.23.473862 & https://doi.org/10.1038/s42003-022-04040-1
W. C. Kao, C. Ortmann de Percin Northumberland, T. C. Cheng, J. Ortiz, A. Durand, O. von Loeffelholz, O. Schilling, M. L. Biniossek, B. P. Klaholz, C. Hunte. Structural basis for safe and efficient energy conversion in a respiratory supercomplex. Nat Commun2022, 13, 545. https://doi.org/10.1038/s41467-022-28179-x.
B. P. Klaholz. Studying the Structural Organization of Polyribosomes with Alexander S. Spirin. Biochemistry (Moscow), 2021, 86, 1053-1059; invited review. https://doi.org/10.1134/S0006297921090030.
A. Ibrahim, C. Papin, K. Mohideen-Abdul, S. Le Gras, I. Stoll, C. Bronner, S. Dimitrov, B. P. Klaholz & A. Hamiche. MeCP2 is a microsatellite binding protein that protects CA repeats from nucleosome invasion. Science2021; 372, 6549, eabd5581. https://doi.org/10.1126/science.abd5581.
O. von Loeffelholz, B. P. Klaholz. Setup and troubleshooting of Volta phase plate cryo-EM data collection. 3rd Edition Methods in Molecular Biology - Structural Proteomics, 2021, 2305, 291-299. https://doi.org/10.1007/978-1-0716-1406-8_14.
D. Rovito, A. I. Rerra, V. Ueberschlag-Pitiot, S. Joshi, N. Karasu, V. Dacleu-Siewe, K. B. Rayana, K. Ghaibour, M. Parisotto, A. Ferry, S. A. Jelinsky, G. Laverny, B. P. Klaholz, T. Sexton, I. M. L. Billas, D. Duteil, D. Metzger. Myod1 and GR coordinate myofiber-specific transcriptional enhancers. Nucleic Acids Res.2021, gkab226. https://doi.org/10.1093/nar/gkab226.
J. Wang, S. K. Natchiar, P. B. Moore& B. P. Klaholz. Identification of Mg2+ ions next to nucleotides in cryo-EM maps using electrostatic potential maps. Acta Cryst D. 2021, D77, 534-539. https://doi.org/10.1107/S2059798321001893.
L. Andronov, J.-L. Vonesch & B. P. Klaholz. Practical aspects of super-resolution imaging and segmentation of macromolecular complexes by dSTORM. Methods Mol Biol.2021, 2247:271-286. https://doi.org/10.1007/978-1-0716-1126-5_15. Invited Book chapter.
Beinsteiner B, Markov GV, Erb S, Chebaro Y, McEwen AG, Cianferani S, Moras D, Laudet V, Billas IML. A Structural Signature Motif Enlightens the Origin and Diversification of Nuclear Receptors, PLoS Genet. 2021, 17, e1009492. http://dx.doi.org/10.1371/journal.pgen.1009492.
Browning C., McEwen A.G., Mori K., Yokoi T., Moras D., Nakagawa Y., Billas I.M.L. Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP. J Pestic Sci.2021 Feb 20;46(1):88-100. http://dx.doi.org/10.1584/jpestics.D20-095.
V. Marcel, J. Kielbassa, V. Marchand, K. S. Natchiar, H. Paraqindes, F. Nguyen van Long, L. Ayadi, V. Bourguignon-Igel, P. Lo Monaco, D. Monchiet, V. Scott, L. Tonon, S. E. Bray, A. Diot, L. B. Jordan, A. M. Thompson, J-C. Bourdon, T. Dubois, F. André, F. Catez, A. Puisieux, Y. Motorin, B. P. Klaholz, A. Viari, J-J. Diaz. Ribosomal RNA 2′O-methylation as a novel layer of inter-tumour heterogeneity in breast cancer. NAR Cancer, 2020, 2, 4. https://doi.org/10.1093/narcan/zcaa036.
I. Orlov, C. Hemmer, L. Ackerer, B. Lorber, A. Ghannam, V. Poignavent, K. Hleibieh, C. Sauter, C. Schmitt-Keichinger, L. Belval, J-M. Hily, A. Marmonier, V. Komar, S. Gersch, P. Schellenberger, P. Bron, E. Vigne, S. Muyldermans, O. Lemaire, G. Demangeat, C. Ritzenthaler & B. P. Klaholz. Structural basis of nanobody-recognition of grapevine fanleaf virus and of virus resistance loss. PNAS, 2020, 117, 10848-10855. http://biorxiv.org/cgi/content/short/728907v1 and https://doi.org/10.1073/pnas.1913681117. Epub 2020 May 5.
V. Bhaskar, A. Graff-Meyer, A. D. Schenk, S. Cavadini, O. von Loeffelholz, S. K. Natchiar, C. G. Artus-Revel, H-R. Hotz, G. Bretones, B. P. Klaholz & J. A. Chao. Dynamics of uS19 C-terminal tail during the translation elongation cycle in human ribosomes. Cell Reports, 2020, 31, 107473. http://doi.org/10.1016/j.celrep.2020.03.037.
A. Gilles, L. Fréchin, S. K. Natchiar, G. Biondani, O. von Loeffelholz, S. Holvec, J-L. Malaval, J-Y. Winum, B. P. Klaholz, J-F. Peyron. Targeting the Human 80S Ribosome in Cancer: From Structure to Function and Drug Design for Innovative Adjuvant Therapeutic Strategies. Cells, 2020, 9 (3). pii: E629; invited review. http://doi.org/10.3390/cells9030629.
Liebschner, D., Urzhumtsev, A., Adams, P.D. & Afonine, P.V. Implementation of riding hydrogen model in CCTBX to support new generaton of X-ray and neutron joint refinement in Phenix. In Methods in Enzymology, Academic Press, San Diego, P.C.E.Moody, ed., 2020, 634, 177-199. http://dx.doi.org/10.1016/bs.mie.2020.01.007.
Urzhumtsev, A. & Lunin, V.Y. Introduction to crystallographic refinement of macromolecular atomic models. Addendum. Cryst. Rev.2020, 26, 51-55. https://doi.org/10.1080/0889311X.2019.1698032.
Zheng, M., Biczysko, M., Xu, Y., Moriarty, N.W., Kruse, H., Urzhumtsev, A., Waller, M.P., Afonine, P.V. Including Crystallographic Symmetry in Quantum-based Refinement: Q|R#2. Acta Cryst.2020, D76, 41-50. http://dx.doi.org/10.1107/S2059798319015122.
B. P. Klaholz. Deriving and refining atomic models in crystallography and cryo-EM: the latest Phenix tools to facilitate structure analysis. Acta Cryst.,2019, D75, 878–881; invited comment. https://doi.org/10.1107/S2059798319013391.
L. Andronov, K. Ouararhni, I. Stoll, B. P. Klaholz & A. Hamiche. CENP-A nucleosome clusters form rosette-like structures around HJURP during G1. Nat Commun., 2019, 10, 4436. https://doi.org/10.1038/s41467-019-12383-3.
C. H. Hill, Boreikaite V, Kumar A, Casanal A, Kubik P, Degliesposti G, Maslen S, Mariani A, von Loeffelholz O, Girbig M, Skehel M, Passmore LA. Activation of the Endonuclease that Defines mRNA 3' Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex. Mol Cell, 2019; 73:1217-1231. https://doi.org/10.1016/j.molcel.2018.12.023.
H. Ma, X. Wang, J. Cai, Q. Dai, S. K. Natchiar, R. Lv, K. Chen, Z. Lu, H. Chen, Y. G. Shi, F. Lan, J. Fan, B. P. Klaholz, T. Pan, Y. Shi, C. He. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol., 2019, 15, 88-94. https://doi.org/10.1038/s41589-018-0184-3.
Urzhumtsev, A. & Lunin, V.Y. Introduction to crystallographic refinement of macromolecular atomic models. Cryst. Rev.2019, 25, 164-262. https://doi.org/10.1080/0889311X.2019.1631817.
Liebschner, D., Afonine, P.V., Baker, M.L., Bunkóczi, G., Chen, V.B., Croll, T.I., Hintze, B.J., Hung,L.-W., Jain, S., McCoy, A.J., Moriarty, N.W., Oeffner, R.D., Poon, B.K., Prisant, M.G., Read, R.J., Richardson, J.S., Richardson, D.C., Sammito, M.D., Sobolev, O.V., Stockwell, D.H., Terwilliger, T.C., Urzhumtsev, A., Videau, L.L., Williams, C.J., Adams, P.D. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Cryst. 2019, D75, 861-877. http://dx.doi.org/10.1107/S2059798319011471.
Urzhumtseva, L., Urzhumtsev, A. py_convrot: rotation conventions, to understand and to apply. J. Appl.Cryst.2019, 52, 869-881. https://doi.org/10.1107/S1600576719007313.
Lunin, V.Y., Lunina, N.L., Petrova, T.E., Baumstark, M.W. & Urzhumtsev, A. Mask-based approach to phasing of single-particle diffraction data. II. Likelihood based selection criteria. Acta Cryst.2019, D75, 79-89. http://dx.doi.org/10.1107/S2059798318016959.
S. K. Natchiar, A. G. Myasnikov, I. Hazemann & B. P. Klaholz. Visualizing the role of 2’-OH rRNA methylations in the human ribosome structure. Biomolecules, 2018, 8; special issue on rRNA biology; invited review. https://doi.org/10.3390/biom8040125.
J. D. Mortison, M. Schenone, J. A. Myers, Z. Zhang, L. Chen, C. Ciarlo, E. Comer, S. K. Natchiar, S. A. Carr, B. P. Klaholz, A. G. Myers. Tetracyclines modify translation by targeting key human rRNA substructures. Cell Chem Biol., 2018, Oct 5. https://doi.org/10.1016/j.chembiol.2018.09.010 and https://www.biorxiv.org/content/early/2018/02/01/256230.
P. V. Afonine, B. P. Klaholz, N. W. Moriarty, B. K. Poon, O. V. Sobolev, T. C. Terwilliger, P. D. Adams & A. Urzhumtsev. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Cryst.2018, D74, 814-840. https://doi.org/10.1107/S2059798318009324.
L. Andronov, J. Michalon, K. Ouararhni, I. Orlov, A. Hamiche, J.-L. Vonesch & B. P. Klaholz.3DClusterViSu: 3D clustering analysis of super-resolution microscopy data by 3D Voronoi tessellations. Bioinformatics, 2018, 34, 3004-3012. http://dx.doi.org/10.1093/bioinformatics/bty200 & https://www.biorxiv.org/content/early/2017/06/07/146456.
D. Bonnard, E. Le Rouzic, S. Eiler, C. Amadori, I. Orlov, J.M. Bruneau, J. Brias, J. Barbion, F. Chevreuil, D. Spehner, S. Chasset, B. Ledoussal, F. Moreau, A. Saïb, B. P. Klaholz, S. Emiliani, M. Ruff, A. Zamborlini, R. Benarous. Structure-function analyses unravel distinct effects of allosteric inhibitors of HIV-1 integrase on viral maturation and integration. J. Biol. Chem.,2018, 293, 6172-6186. http://dx.doi.org/10.1074/jbc.M117.816793.
O. von Loeffelholz, G. Papai, R. Danev, A. G. Myasnikov, S. K. Natchiar, I. Hazemann, J.-F. Ménétret & B. P. Klaholz. Volta phase plate data collection facilitates image processing and cryo-EM structure determination. J. Struct. Biol., 2018, 202, 191-199. http://dx.doi.org/10.1016/j.jsb.2018.01.003.
I. Orlov, R. Drillien, D. Spehner, M. Bergoin, A. Abd-Alla, B. P. Klaholz. Structural features of the Salivary gland hypertrophy virus of the tsetse fly revealed by cryo electron microscopy and tomography. Virology, 2018, 514, 165-169. http://dx.doi.org/10.1016/j.virol.2017.11.016.
Khalturin K, Billas IML, Chebaro Y, Reitzel AM, Tarrant AM, Laudet V, Markov GV, 2018, NR3E receptors in cnidarians: A new family of steroid receptor relatives extends the possible mechanisms for ligand binding. J Steroid Biochem Mol Biol.2018, 184, 11-19. http://dx.doi.org/10.1016/j.jsbmb.2018.06.014.
Lunina, N.L., Petrova, T.E., Urzhumtsev, A., Lunin, V.Y. The use of connected masks for reconstructing the single particle images from X-ray diffraction data. III. Maximum-likelihood based strategies to select solution of the Phase problem. Math. Biol. Bioinformatics 2018,13, t70-t83. https://doi.org/10.17537/2018.13.t70.
Afonine, P.V., Adams, P.D. & Urzhumtsev, A. From deep TLS validation to ensembles of atomic models built from elemental motions. II. Analysis of TLS refinement results by explicit interpretation. Acta Cryst.2018,D74, 621-631. http://dx.doi.org/10.1107/S2059798318005764.
Afonine, P.V., Poon, B.K., Read, R.J., Sobolev, O.V., Terwilliger, T.C., Urzhumtsev, A. & Adams, P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Cryst.2018, D74, 531-544. http://dx.doi.org/10.1107/S2059798318006551.
S. K. Natchiar, A. G. Myasnikov, H. Kratzat, I. Hazemann & B. P. Klaholz. Atomic model building and refinement into high-resolution cryo-EM maps. Protocol Exchange,2017http://dx.doi.org/10.1038/protex.2017.122.
S. K. Natchiar, A. G. Myasnikov, H. Kratzat, I. Hazemann & B. P. Klaholz. Visualization of chemical modifications in the human 80S ribosome structure. Nature, 2017, 551, 472-477. http://dx.doi.org/10.1038/nature24482. Epub 2017 Nov 15.
C. Amadori, Y. U. van der Velden, D. Bonnard, I. Orlov, N. van Bel, E. Le Rouzic, L. Miralles, J. Brias, F. Chevreuil, D. Spehner, S. Chasset, B. Ledoussal, L. Mayr, F. Moreau, F. Garcia, J. Gatell, A. Zamborlini, S. Emiliani, M. Ruff, B. P. Klaholz, C. Moog, B. Berkhout, M. Plana & R. Benarous. The HIV-1 integrase-LEDGF allosteric inhibitor MUT-A: resistance profile, impairment of virus maturation and infectivity but without influence on RNA packaging or virus immunoreactivity. Retrovirology,2017, 14, 50. http://dx.doi.org/10.1186/s12977-017-0373-2.
O. von Loeffelholz, S. K. Natchiar, N. Djabeur, A. G. Myasnikov, H. Kratzat, J.-F. Ménétret, I. Hazemann & B. P. Klaholz. Focused classification and refinement in high-resolution cryo-EM structural analysis of ribosome complexes.Curr. Opin. Struct. Biol., 2017, 46, 140-148; invited review. http://dx.doi.org/10.1016/j.sbi.2017.07.007.
B. P. Klaholz. The ribosome holds the RNA Polymerase on Track in Bacteria. Trends Biochem Sci., 2017, 42, 686-689; invited review. http://dx.doi.org/10.1016/j.tibs.2017.07.003.
K. Mohideen-Abdul, K. Tazibt, M. Bourguet M, I. Hazemann, I. Lebars, M. Takacs, S. Cianférani, B. P. Klaholz, D. Moras & I. M. L. Billas. Importance of the Sequence-Directed DNA Shape for Specific Binding Site Recognition by the Estrogen-Related Receptor. Front. Endocrinol.2017, 8, 140. http://dx.doi.org/10.3389/fendo.2017.00140.
I. Orlov, A. G. Myasnikov, L. Andronov, S. K. Natchiar, H. Khatter, B. Beinsteiner, J-F. Ménétret, I. Hazemann, K. Mohideen, K. Tazibt, R. Tabaroni, H. Kratzat, N. Djabeur, T. Bruxelles, F. Raivoniaina, L. di Pompeo, M. Torchy, I. Billas, A. Urzhumtsev& B. P. Klaholz. The integrative role of cryo electron microscopy in molecular and cellular structural biology. Biol Cell.,2017, 109, 81-93; invited review. http://dx.doi.org/10.1111/boc.201600042.
Markov GV, Gutierrez-Mazariegos J, Pitrat D, Billas IML, Bonneton F, Moras D, Hasserodt J, Lecointre G, Laudet V (2017) Origin of an ancient hormone/receptor couple revealed by resurrection of an ancestral estrogen. Sci Adv. 2017, 3, e1601778. http://dx.doi.org/10.1126/sciadv.1601778.
Carnesecchi J, Forcet C, Zhang L, Tribollet V, Barenton B, Boudra R, Cerutti C, Billas I.M.L, Sérandour A, Carroll J, Beaudoin C, Vanacker J-M. ERRα induces H3K9 demethylation by LSD1 to promote cell invasion. PNAS2017, 114, 3909-3914. http://dx.doi.org/10.1038/s41598-018-27676-8.
Aptekarev, A.I., Afendikov, A.L., Ataullkhanov, F.I., Balabaev, N.K., Biktashev, V.N., Biktasheva, I.V., Borisyuk, R.M., Vvedenskaya, N.D., Dagkesamansky, R.D., Zarkhin, Yu.G., Ilyashenko, Yu.S., Lakhno, V.D., Lunin, V.Yu., Lunina, N.L., Nikolaev, E.V., Posvyansky, V.S., Roytberg, M.A., Ryabenky, V.S., Ryashko, L.B., Sinay, Ya.G., Tikhomirov, V.M., Tokarev, A.A., Urzhumtsev, A.G. & Khibnik, A.I. () "In memory of Emmanuil Elievich Shnol". Russian Mathematical Surveys (Uspekhi Matematicheskikh Nauk, in russ.)2017, 72, 197-208. http://dx.doi.org/10.1070/RM9751.
P.-D. Coureux, C. Lazennec-Schurdevin, A. Monestier, E. Larquet, L. Cladière, B. P. Klaholz, E. Schmitt & Y. Mechulam. Initiator tRNA accommodation during translation initiation mRNA probing. Cryo-EM study of start codon selection during archaeal translation initiation. Nat. Commun., 2016, 7, 13366. http://dx.doi.org/10.1038/ncomms13366.
A. G. Myasnikov, S. K. Natchiar, M. Nebout, I. Hazemann, V. Imbert, H. Khatter, J.-F. Peyron & B. P. Klaholz. Structure-function insights reveal the human ribosome as a cancer target for antibiotics. Nat. Commun., 2016, 7, 12856. http://dx.doi.org/10.1038/ncomms12856.
F. Martin, J.-F. Ménétret, A. Simonetti, A. G. Myasnikov, Q. Vicens, L. Prongidi-Fix, S. K. Natchiar, B. P. Klaholz & G. Eriani. Ribosomal 18S rRNA base pairs with mRNA during eukaryotic translation initiation. Nat. Commun., 2016, 7, 12622. http://dx.doi.org/10.1038/ncomms12622.
L. Andronov, I. Orlov, Y. Lutz, J-L. Vonesch & B. P. Klaholz. ClusterViSu, a method for clustering of protein complexes by Voronoi tessellation in super-resolution microscopy. Sci. Rep.2016, 6, 24084. http://dx.doi.org/10.1038/srep24084.
L. Andronov, Y. Lutz, J-L. Vonesch & B. P. Klaholz. SharpViSu: integrated analysis and segmentation of super-resolution microscopy data. Bioinformatics, 2016, 32, 2239-2241. http://dx.doi.org/10.1093/bioinformatics/btw123.
S. Thebault, M. Agez, X. Chi, J. Stojko, V. Cura, S.B. Telerman, L. Maillet, F. Gautier, I.M.L. Billas-Massobrio, C. Birck, N. Troffer-Charlier, T. Karafin, J. Honore, A. Senff-Ribeiro, S. Montessuit, C.M. Johnson, P. Juin, S. Cianferani, J.C. Martinou, D.W. Andrews, R. Amson, A. Telerman, J. Cavarelli. TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL. Sci. Rep.2016, 6, 19725. http://dx.doi.org/10.1038/srep19725.
Urzhumtsev, A., Urzhumtseva, L., Baumann, U. Helical symmetry of nucleic acids: obstacle or help? In: "Nucleic Acids Crystallography: Methods and Protocols" (Springer Protocols), E.Ennifar, ed., New York – Heidelberg-Dordrecht-London: Springer (Humana Press), 2016, 259-267. http://dx.doi.org/10.1007/978-1-4939-2763-0_16.
Urzhumtseva, L., Urzhumtsev, A. COMPaRS: stand-alone program for map comparison using quantile rang scaling. J. Appl.Cryst.2016, 49, 2270-2275. https://doi.org/10.1107/S1600576716015752.
Urzhumtsev, A., Afonine, P.V., Van Benschoten, A.H., Fraser, J.S. & Adams, P.D. From deep TLS validation to ensembles of atomic models built from elemental motions. Addenda and corrigendum. Acta Cryst.2016, D72, 1073-1075. http://dx.doi.org/10.1107/S2059798316013048.
Lunin, V.Y., Lunina, N.L., Petrova, T.E., Baumstark, M.W. & Urzhumtsev, A. Mask-based approach to phasing of single-particle diffraction data. Acta Cryst2016, D72, 147-157. http://dx.doi.org/10.1107/S2059798318016959.
B. P. Klaholz. Structure sorting of multiple macromolecular states in heterogeneous cryo-EM samples by 3D multivariate statistical analysis. Open Journal of Statistics, Special Issue on Multivariate Statistical Analysis, 2015, 5, 820-836. http://dx.doi.org/10.4236/ojs.2015.57081.
V. Postupalenko, D. Desplancq, I. Orlov, Y. Arntz, D. Spehner, Y. Mely, B. P. Klaholz, P. Schultz, E. Weiss, G. Zuber. Protein Delivery System Containing a Nickel-Immobilized Polymer for Multimerization of Affinity-Purified His-Tagged Proteins Enhances Cytosolic Transfer. Angew Chem Int Ed Engl., 2015. http://dx.doi.org/10.1002/anie.201505437.
B. Beinsteiner, J. Michalon & B. P. Klaholz. IBiSS, a versatile and interactive tool for integrated sequence and 3D structure analysis of large macromolecular complexes. Bioinformatics, 2015, 31, 3339-3344. http://dx.doi.org/10.1093/bioinformatics/btv347.
S. K. Natchiar, H. Khatter, A. G. Myasnikov, & B. P. Klaholz. Structure of the human 80S ribosome. Acta Cryst., 2015, A71, s33. http://dx.doi.org/10.1107/s2053273315099477.
H. Khatter, A. G. Myasnikov, K. Natchiar & B. P. Klaholz. Structure of the human 80S ribosome. Nature, 2015, 520, 640-5. http://dx.doi.org/10.1038/nature14427. Epub 2015 Apr 22.
Moras, I. Billas, N. Rochel, B. P. Klaholz. Structure–function relationships in nuclear receptors: the facts. Trends Biochem Sci., 2015, 1136 1-4. http://dx.doi.org/10.1016/j.tibs.2015.03.009.
P. Leone, C. Bebeacua, O. Opota, C. Kellenberger, B. P. Klaholz, I. Orlov, C. Cambillau, B. Lemaitre, A. Roussel. X-ray and Cryo-electron Microscopy Structures of Monalysin Pore Forming Toxin Reveals Multimerization of the Pro-Form. J. Biol. Chem., 2015 Apr 5. pii: jbc.M115.646109. http://dx.doi.org/10.1074/jbc.M115.646109.
M. P. Torchy, A. Hamiche & B. P. Klaholz. Structure and function insights into the NuRD chromatin-remodeling complex. Cell. Mol. Life Sci., 2015, 72, 2491–2507. http://dx.doi.org/10.1007/s00018-015-1880-8.
E. Uchikawa, K. S. Natchiar, X. Han, F. Proux, P. Roblin, E. Zhang, A. Durand, B. P. Klaholz, A. C. Dock-Bregeon. Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LaRP7. Nucleic Acids Res.2015, 43, 3373-88. http://dx.doi.org/10.1093/nar/gkv173.
H. Anton, N. Taha, E. Boutant, L. Richert, H. Khatter, B. P. Klaholz, P. Rondé, E. Réal, H. de Rocquigny & Y. Mély. Investigating the cellular distribution and interactions of HIV-1 Nucleocapsid protein by quantitative fluorescence microscopy. PlosOne2015, 10, e0116921. http://dx.doi.org/10.1371/journal.pone.0116921. eCollection 2015.
I. Orlov, A. Schertel, G. Zuber, B. P. Klaholz, R. Drillien, E. Weiss, P. Schultz & D. Spehner. Live cell immunogold labelling of RNA polymerase II. Sci. Rep.2015, 5, 8324. http://dx.doi.org/10.1038/srep08324.
Z. A. Afonina, A. G. Myasnikov, V. A. Shirokov, B. P. Klaholz, A. S. Spirin. Conformation transitions of eukaryotic polyribosomes during multi-round translation. Nucleic Acids Res.2015, 43, 618-28. http://dx.doi.org/10.1093/nar/gku1270.
A. G. Myasnikov, Z. A. Afonina, J-F. Ménétret, V. A. Shirokov, A. S. Spirin & B. P. Klaholz. The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes. Nat. Commun., 2014, 5, 5294. http://dx.doi.org/10.1038/ncomms6294.
C. Lemaître, A. Grabarz, K. Tsouroula, L. Andronov, A. Furst, T. Pankotai, V. Heyer, M. Rogier, K.M. Attwood, P. Kessler, G. Dellaire, B. Klaholz, B. Reina-San-Martin, E. Soutoglou. Nuclear position dictates DNA repair pathway choice. Genes Dev.2014, pii: gad.248369.114; http://dx.doi.org/10.1101/gad.248369.114.
Z. A. Afonina, A. G. Myasnikov, V. A. Shirokov, B. P. Klaholz, A. S. Spirin. Formation of circular polyribosomes on eukaryotic mRNA without cap-structure and poly(A)-tail: a cryo electron tomography study. Nucleic Acids Res.,2014, 42, 9461-9. http://dx.doi.org/10.1093/nar/gku599.
M. Maletta, I. Orlov, P. Roblin, Y. Beck, D. Moras, I. M. L. Billas & B. P. Klaholz. The palindromic DNA-bound USP/EcR nuclear receptor adopts an asymmetric organization with allosteric domain positioning. Nat. Commun., 2014, 5, 4139. http://dx.doi.org/10.1038/ncomms5139.
B. P. Klaholz. Microscopy and Microanalysis, 2014, 20, 991-991. http://dx.doi.org/10.1017/S1431927614000725. Invited book review.
S. Spinelli, C. Bebeacua, I. Orlov, D. Tremblay, B. P. Klaholz, S. Moineau, C. Cambillau. CryoEM structure of the lactococcal siphophage 1358 virion. J Virol.2014. pii: JVI.01040-14; http://dx.doi.org/10.1128/JVI.01040-14.
H. Khatter, A. G. Myasnikov, L. Mastio, I. M. L. Billas, C. Birck, S. Stella & B. P. Klaholz. Purification, characterization and crystallization of the human 80S ribosome. Nucleic Acids Res., 2014, 42(6), 1-11; http://dx.doi.org/10.1093/nar/gkt1404.
M. Duval, A. Korepanov, O. Fuchsbauer, P. Fechter, A. Haller, A. Fabbretti, L. Choulier, R. Micura, B. P. Klaholz, P. Romby, M. Springer & S. Marzi. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLOS Biol., 2013, 11 (12):e1001731; http://dx.doi.org/10.1371/journal.pbio.1001731.
A. Simonetti, S. Marzi, I. M. L. Billas, A. Tsai, A. Fabbretti, A. G. Myasnikov, P. Roblin, A. C. Vaiana, I. Hazemann, D. Eiler, T. A. Steitz, J. D. Puglisi, C. O. Gualerzi & B. P. Klaholz. Involvement of IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc. Nat. Acad. Sci., 2013, 110, 15656-15661; http://dx.doi.org/10.1073/pnas.1309578110.
D. Eiler, J. Lin, A. Simonetti, B. P. Klaholz & T. A. Steitz. IF2 Initiation factor 2 crystal structure reveals a different domain organization from eukaryotic initiation factor 5B and mechanism among translational GTPases. Proc. Nat. Acad. Sci., 2013, 110, 15662-15667; http://dx.doi.org/10.1073/pnas.1309360110.
L. Urzhumtseva, B. P. Klaholz & A. Urzhumtsev. On effective and optical resolutions of diffraction data sets. Acta Cryst., 2013, D69, 1921-1934; http://dx.doi.org/10.1107/S0907444913016673.
J-F. Ménétret, H. Khatter, A. Simonetti, I. Orlov, A. G. Myasnikov, Vidhya KV, S. Manicka, M. Torchy, K. Mohideen, A-S. Humm, I. Hazemann, A. Urzhumtsev, B. P. Klaholz. Integrative structure-function analysis of large nucleoprotein complexes. RNA Structure and Folding, de Gruyter, 2013, D. Klostermeier & C. Hammann (Eds.); invited review/book chapter. https://doi.org/10.1515/9783110284959.
A. Simonetti, S. Marzi, A. Fabbretti, I. Hazemann, L. Jenner, A. Urzhumtsev, C. O. Gualerzi & B. P. Klaholz. Structure of the protein core of translation initiation factor 2 in apo, GTP-bound and GDP-bound forms. Acta Cryst., 2013, D69, 925-933; http://dx.doi.org/10.1107/S0907444913006422.
Z. A. Afonina, A. G. Myasnikov, N .F. Khabibullina, A. Y. Belorusova, J-F. Ménétret, V. D. Vasiliev, B. P. Klaholz, V. A. Shirokov, A. S. Spirin. Topology of mRNA chain in isolated eukaryotic double-row polyribosomes. Biochemistry (Mosc), 2013, 78, 445-54; http://dx.doi.org/10.1134/S0006297913050027.
A. G. Myasnikov, Z. Afonina, B. P. Klaholz. Single particle and molecular assembly analysis of polyribosomes by single- and double-tilt cryo electron tomography. Ultramicroscopy, 2013, 126, 33-39; http://dx.doi.org/10.1016/j.ultramic.2012.12.009.
L. Prongidi-Fix, L. Schaeffer, A. Simonetti, S. Barends, J-F. Ménétret, B. P. Klaholz, G. Eriani, F. Martin. Rapid purification of ribosomal particles assembled on histone H4 mRNA: a new method based on mRNA-DNA chimaeras. Biochem J.2013, 449, 719-28; http://dx.doi.org/10.1042/BJ20121211.
L. Amoasii, D. L. Bertazzi, H. Tronchère, K. Hnia, G. Chicanne, B. Rinaldi, B. S. Cowling, A. Ferry, B. P. Klaholz, B. Payrastre, J. Laporte, S. Friant. Phosphatase-dead myotubularin ameliorates X-linked centronuclear myopathy phenotypes in mice. PLoS Genet.2012, 8(10):e1002965; http://dx.doi.org/10.1371/journal.pgen.1002965.
I. Orlov, N. Rochel, D. Moras & B. P. Klaholz. Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J., 2012, 31, 291-300; http://dx.doi.org/10.1038/emboj.2011.445.
A. Simonetti, S. Marzi, A. G. Myasnikov, J-F. Ménétret & B. P. Klaholz. Insights into translation initiation and termination complexes and into the polysome architecture. Ribosomes; Rodnina/Wintermeyer/Green (Eds), Springer, 2011; invited review/book chapter. http://dx.doi.org/10.1007/978-3-7091-0215-2_10.
B. P. Klaholz. Molecular recognition and catalysis in translation termination complexes. Trends Biochem. Sci., 2011, 36, 282-292; http://dx.doi.org/10.1016/j.tibs.2011.02.001; invited review.
B. P. Klaholz. Let's see how tmRNA rescues a stuck ribosome. EMBO J., 2010, 17, 3747-9; http://dx.doi.org/10.1038/emboj.2010.269; invited highlight/review.
A. G. Myasnikov, Z. Afonina, A. Simonetti, S. Marzi, B. P. Klaholz. Investigation of the protein synthesis machinery at different levels of organization through integrative cryo-EM. Acta Cryst. A66, 2010; s21-s21. http://dx.doi.org/10.1107/S0108767310099538.
A. G. Myasnikov, A. Simonetti, S. Marzi, B. P. Klaholz. Structure-function insights into prokaryotic and eukaryotic translation initiation.Curr. Op. Struct. Biol., 2009, 19, 300-309; invited review http://dx.doi.org/10.1016/j.sbi.2009.04.010.
J-F. Ménétret & B. P. Klaholz. La synthèse des protéines vue au cryomicroscope électronique [Protein synthesis seen under the electron microscope]. Biofutur, 2009, 305, 50-53; invited review.
A. Simonetti, S. Marzi, L. Jenner, A. Myasnikov, P. Romby, G. Yusupova, B. P. Klaholz & M. Yusupov. A structural view of translation initiation in bacteria. Cell. Mol. Life Sci., 2009, 66, 423-436; invited review. http://dx.doi.org/10.1007/s00018-008-8416-4.
A. Simonetti, S. Marzi, A. G. Myasnikov, A. Fabbretti, M. Yusupov, C. O. Gualerzi & B. P. Klaholz. Purification of T. thermophilus translation initiation factors and characterization of the corresponding ribosome complexes by filter-binding assays. Protocol Exchange,2008.http://dx.doi.org/10.1038/nprot.2008.130.
A. Simonetti, S. Marzi, A. G. Myasnikov, A. Fabbretti, G. Yusupova, M. Yusupov, C. O. Gualerzi & B. P. Klaholz. Structure of the 30S translation initiation complex. Nature, 2008, 455, 416-420. http://dx.doi.org/10.1038/nature07192.
S. Marzi, P. Romby & B. P. Klaholz. Quand le fil de l’ARN messager s’entortille et bloque sa traduction (Twisting of mRNA reversibly blocks its translation by the ribosome). Med. & Sci. (Paris). 2007, 23, 881-884; review.
S. Marzi, A. G. Myasnikov, A. Serganov, C. Ehresmann, P. Romby, M. Yusupov & B. P. Klaholz. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell, 2007, 130, 1019–1031. http://dx.doi.org/10.1016/j.cell.2007.07.008.
A. G. Myasnikov, S. Marzi, A. Simonetti, A. M. Giuliodori, C. O. Gualerzi, G. Yusupova, M. Yusupov & B. P. Klaholz. Conformational transition of initiation factor 2from the GTP- to GDP-bound state visualized on the ribosome. Nat. Struct. Mol. Biol., 2005, 12, 1145-1149. http://dx.doi.org/10.1038/nsmb1012.
B. P. Klaholz, A. G. Myasnikov & M. van Heel. Visualisation of release factor 3 on the ribosome during termination of protein synthesis. Nature, 2004, 427, 862-865. http://dx.doi.org/10.1038/nature02332.
P. Ray, B. P. Klaholz, R. D. Finn, E. V. Orlova, P. C. Burrows, B. Gowen, M. Buck & M. van Heel. Determination of Escherichia coli RNA polymerase structure by single particle cryo-electron microscopy. Methods in Enzymology, 2003, 370, 24-42. https://doi.org/10.1016/S0076-6879(03)70003-2.
B. P. Klaholz, T. Pape, A. V. Zavialov, A. G. Myasnikov, B. Vestergaard, E. Orlova, M. Ehrenberg & M. van Heel. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature2003, 421, 90-94. http://dx.doi.org/10.1038/nature01225.
News
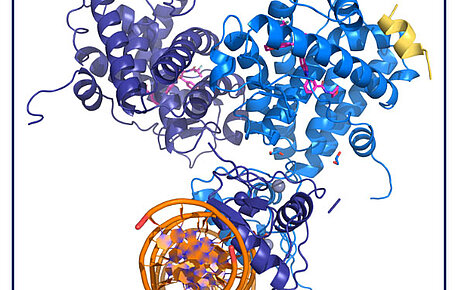
The quaternary structure of the GR highlights inter-domain allosteric communication
In inflammatory or autoimmune diseases, modulating the action of the glucocorticoid receptor (GR) is an effective therapeutic strategy. However, the…
Read more
Awards and recognitions
Samuel Holvec - PhD presentation prize of the RNA Society at the EpiRNA meeting - 2023
Stefano Stella - Knight of the Order of Merit of the Italian Republic – 2022
Leonid Andronov - PhD thesis prize of the doctoral school – 2019
Bruno Klaholz - Médaille d'Argent - CNRS – 2018
Bruno Klaholz - Fellow of the University of Strasbourg Institute of Advanced Studies (USIAS) – 2018
Bruno Klaholz - Raimond Castaing Prize - French Society of Microscopies – 2017
Bruno Klaholz - Richard Lounsbery prize - Académie des sciences – 2016
Bruno Klaholz - ERC Starting grant - European Research Council (ERC) – 2009
Bruno Klaholz - Nuclear Receptor Conference Presentation Prize - European Molecular Biology Organization (EMBO) – 2009
Bruno Klaholz - Bronze Medal - CNRS – 2008
Bruno Klaholz - Paul Mandel Gutenberg Group Prize - CNRS – 2008
Bruno Klaholz - EMBO Young Investigator Award - European Molecular Biology Organization Young Investigator Programme (EMBO YIP) – 2006
Funding and partners
FRM, INCa, ANR, Ligue, ARC, USIAS, IMCBio
