Nuclear organisation and division
Nuclear organisation and division
Living cells have a remarkable ability to organize their internal components in a precise and dynamic manner. This is especially evident during cell division, when cells rapidly reorganize, duplicate their contents, and split into two daughters that often adopt distinct fates. These complex transformations must be tightly coordinated in space and time.
Our lab studies the molecular mechanisms that ensure this coordination, with a focus on three interconnected areas:
First, we investigate how chromosome segregation and cytokinesis are coupled, particularly through the Aurora B-dependent NoCut checkpoint, which prevents abscission in the presence of chromatin bridges.
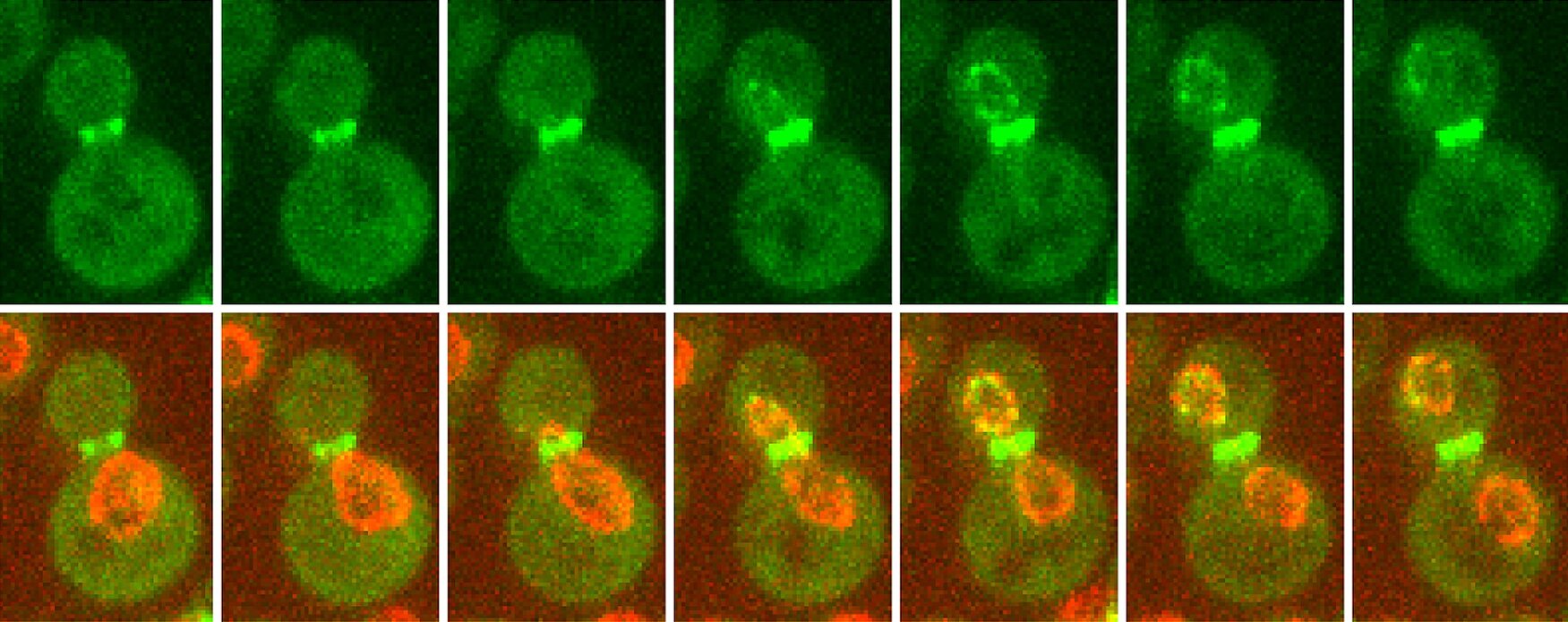
Second, we explore how nuclear organization influences cell identity. We found that nuclear pore complexes (NPCs) are differentially acetylated in mother and daughter cells in budding yeast, and that this modification regulates gene expression and mRNA export. We are now investigating how NPC acetylation contributes to transcriptional and post-transcriptional control, and whether similar regulatory principles apply in mouse embryonic stem (ES) cells. In this context, we focus on the role of non-histone protein acetylation in controlling mRNA processing, export, and stability during cell fate transitions.
Third, we study how cells maintain nuclear homeostasis during aging. In budding yeast, DNA damage in the ribosomal DNA (rDNA) region leads to the formation of extra-chromosomal rDNA circles (ERCs), which accumulate in aging mother cells. We are developing a proximity-labeling proteomics approach to identify proteins that associate with the rDNA during damage and ERC formation. This project, which combines mass spectrometry with live-cell imaging and microfluidics, aims to uncover how rDNA instability impacts nuclear function and how ERCs contribute to aging-related decline in nuclear organization.
Together, our work aims to uncover how cells preserve nuclear integrity and regulate gene expression through dynamic changes in nuclear architecture, with implications for understanding both development and aging.
Members
Researchers
Post-doctoral fellows
PhD students
Engineers
Former members
- Gabriel Neurohr (Group Leader at ETH Zurich)
- Iris Titos (Assistant Professor, Northwestern University, Chicago, Illinois, United States)
- Nuno Amaral (Technical Writer, Sweden)
- Aina Masgrau (Senior Validation Consultant, Spain)
- Andrea Battola (Medical Affairs Manager, Italy)
- Michael Maier (Postdoctoral Researcher, Institute of Medical Biology, Singapore)
- Francesca Di Giovanni (Technical Support Scientist, France)
- Tsvetomira Ivanova
- Arun Kumar (Postdoctoral Researcher, Cell and Molecular Biology Program, Pompeu Fabra University, Spain)
- Petra Stockinger (Research and Development Specialist, Spain)
- Zhanna Shcheprova (Senior Data Team Lead, France)
- Anne Daulny
- Trinidad Sanmartin (laboratory technician, Institute for Bioengineering of Catalonia, Spain)
- Mercè Gomar (Researcher Juan de la Cierva, University of Valencia, Spain)
- Céline Birling (ingénieur d'études, France)
- Vasilisa Pozharskaia (postdoctoral fellow in Université de Laval, Quebec City, Canada)
- Monica Dam (Research Scientist at Skyhawk Therapeutics, Basel, Switzerland)
Current projects
1. Checkpoint control of cytokinesis in response to chromatin bridges
2. Regulation of gene expression and mRNA export by nuclear pore acetylation
3. Mechanisms of rDNA instability and extra-chromosomal rDNA circle accumulation during aging
Funding and partners
- French National Research Agency (ANR), Coordinator of Collaborative Research Project, 2022-2025
- ARC Foundation for Cancer Research, « Programme Labellisé », 2022-2025
- FRM Medical Research Foundation, “Equipe Labellisée”, 2021-2024
Awards and recognitions
European Research Council, Starting Grant (2011-2017)
Publications
-
2025
-
Gene-specific transcript buffering revealed by perturbation of coactivator complexes
- Faezeh Forouzanfar
- David F Moreno
- Damien Plassard
- Audrey Furst
- Karen Amaral de Oliveira
- Bernardo Reina San Martin
- Làszlo Tora
- Nacho Molina
- Manuel Mendoza
Science Advances ; Volume: 11 ; Page: eadr1492
-
-
2024
-
Chromatin Compaction Follows a Power Law Scaling with Cell Size from Interphase Through Mitosis
- Petra Stockinger
- Anna Oddone
- Melike Lakadamyali
- Manuel Mendoza
- Jérôme Solon
BioRxiv
-
Srs2/PARI DNA helicase mediates abscission inhibition in response to chromatin bridges in yeast and human cells
- Monica Dam
- Nicola Brownlow
- Audrey Furst
- Coralie Spiegelhalter
- Manuel Mendoza
BioRxiv
-
Gene-specific RNA homeostasis revealed by perturbation of the NuA4/Tip60 acetyltransferase complex
- Faezeh Forouzanfar
- Damien Plassard
- Damien Plassard
- Audrey Furst
- David F Moreno
- Karen A Oliveira
- Bernardo Reina-San-Martin
- László Tora
- Nacho Molina
- Manuel Mendoza
-
-
2022
-
Nuclear pore complex acetylation regulates mRNA export and cell cycle commitment in budding yeast
- Mercè Gomar-Alba
- Vasilisa Pozharskaia
- Bogdan Cichocki
- Celia Schaal
- Arun Kumar
- Basile Jacquel
- Gilles Charvin
- J Carlos Igual
- Manuel Mendoza
EMBO Journal
-
-
2020
-
The budding yeast Start repressor Whi7 differs in regulation from Whi5, emerging as a major cell cycle brake in response to stress
- Ester Méndez
- Mercè Gomar-Alba
- M. Carmen Bañó
- Manuel Mendoza
- Inma Quilis
- J. Carlos Igual
Journal of Cell Science ; Volume: 133
-
Budding yeast complete DNA synthesis after chromosome segregation begins
- Tsvetomira Ivanova
- Michael Maier
- Alsu Missarova
- Céline Ziegler-Birling
- Monica Dam
- Mercè Gomar-Alba
- Lucas Carey
- Manuel Mendoza
Nature Communications ; Volume: 11
-
Impact of Chromosome Fusions on 3D Genome Organization and Gene Expression in Budding Yeast
- Marco Di Stefano
- Francesca Di Giovanni
- Vasilisa Pozharskaia
- Mercè Gomar-Alba
- Davide Baù
- Lucas Carey
- Marc Marti-Renom
- Manuel Mendoza
Genetics ; Volume: 214 ; Page: 651-667
-
Modulation of Cell Identity by Modification of Nuclear Pore Complexes
- Mercè Gomar-Alba
- Manuel Mendoza
Frontiers in Genetics ; Volume: 10 ; Page: 1301
-
-
2018
-
Daughter-cell-specific modulation of nuclear pore complexes controls cell cycle entry during asymmetric division
- Arun Kumar
- Priyanka Sharma
- Mercè Gomar-Alba
- Zhanna Shcheprova
- Anne Daulny
- Trinidad Sanmartín
- Irene Matucci
- Charlotta Funaya
- Miguel Beato
- Manuel Mendoza
Nature Cell Biology ; Volume: 20 ; Page: 432-442
-

