Nuclear organisation and division
Nuclear organisation and division
Living cells have a remarkable ability to organize their internal components in a precise and dynamic manner. This is especially evident during cell division, when cells rapidly reorganize, duplicate their contents, and split into two daughters that often adopt distinct fates. These complex transformations must be tightly coordinated in space and time.
Our lab studies the molecular mechanisms that ensure this coordination, with a focus on three interconnected areas:
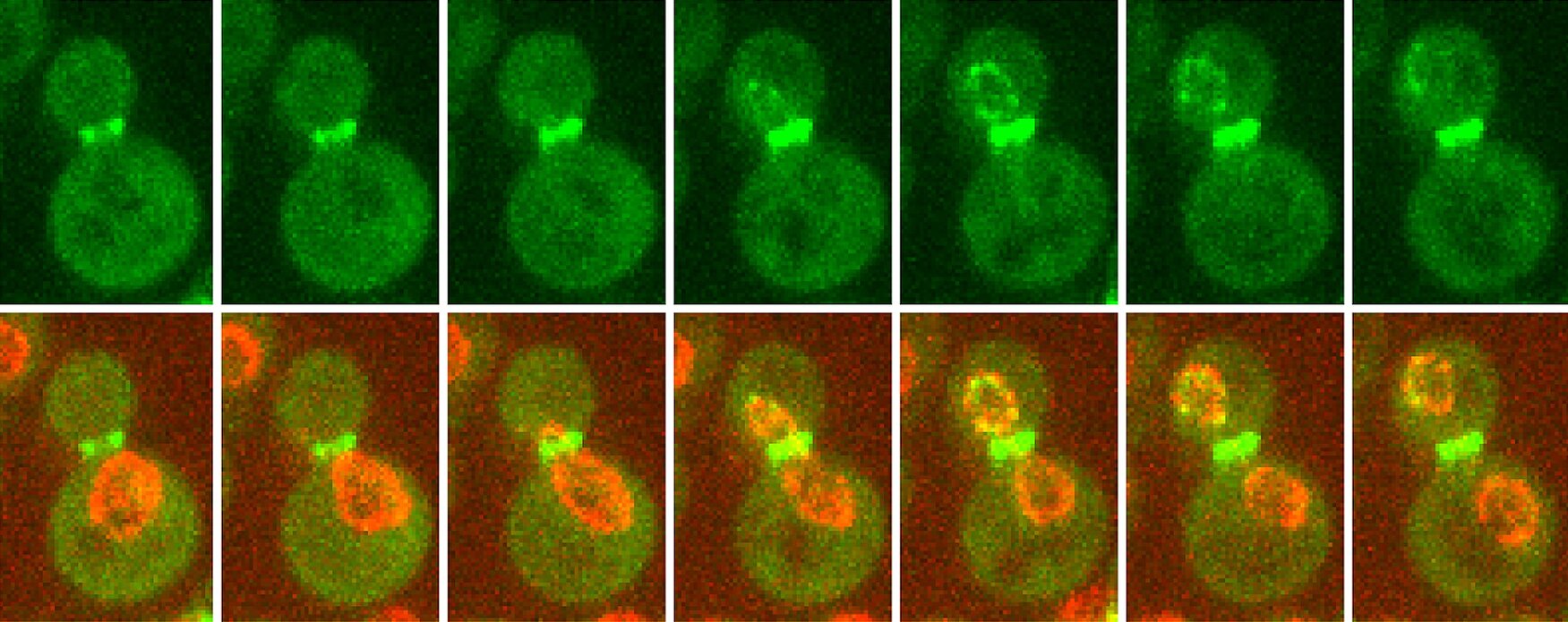
First, we investigate how chromosome segregation and cytokinesis are coupled, particularly through the Aurora B-dependent NoCut checkpoint, which prevents abscission in the presence of chromatin bridges.
Second, we explore how nuclear organization influences cell identity. We found that nuclear pore complexes (NPCs) are differentially acetylated in mother and daughter cells in budding yeast, and that this modification regulates gene expression and mRNA export. We are now investigating how NPC acetylation contributes to transcriptional and post-transcriptional control, and whether similar regulatory principles apply in mouse embryonic stem (ES) cells. In this context, we focus on the role of non-histone protein acetylation in controlling mRNA processing, export, and stability during cell fate transitions.
Third, we study how cells maintain nuclear homeostasis during aging. In budding yeast, DNA damage in the ribosomal DNA (rDNA) region leads to the formation of extra-chromosomal rDNA circles (ERCs), which accumulate in aging mother cells. We are developing a proximity-labeling proteomics approach to identify proteins that associate with the rDNA during damage and ERC formation. This project, which combines mass spectrometry with live-cell imaging and microfluidics, aims to uncover how rDNA instability impacts nuclear function and how ERCs contribute to aging-related decline in nuclear organization.
Together, our work aims to uncover how cells preserve nuclear integrity and regulate gene expression through dynamic changes in nuclear architecture, with implications for understanding both development and aging.
Members
Researchers
Post-doctoral fellows
PhD students
Engineers
Former members
- Gabriel Neurohr (Group Leader at ETH Zurich)
- Iris Titos (Assistant Professor, Northwestern University, Chicago, Illinois, United States)
- Nuno Amaral (Technical Writer, Sweden)
- Aina Masgrau (Senior Validation Consultant, Spain)
- Andrea Battola (Medical Affairs Manager, Italy)
- Michael Maier (Postdoctoral Researcher, Institute of Medical Biology, Singapore)
- Francesca Di Giovanni (Technical Support Scientist, France)
- Tsvetomira Ivanova
- Arun Kumar (Postdoctoral Researcher, Cell and Molecular Biology Program, Pompeu Fabra University, Spain)
- Petra Stockinger (Research and Development Specialist, Spain)
- Zhanna Shcheprova (Senior Data Team Lead, France)
- Anne Daulny
- Trinidad Sanmartin (laboratory technician, Institute for Bioengineering of Catalonia, Spain)
- Mercè Gomar (Researcher Juan de la Cierva, University of Valencia, Spain)
- Céline Birling (ingénieur d'études, France)
- Vasilisa Pozharskaia (postdoctoral fellow in Université de Laval, Quebec City, Canada)
- Monica Dam (Research Scientist at Skyhawk Therapeutics, Basel, Switzerland)
Current projects
1. Checkpoint control of cytokinesis in response to chromatin bridges
2. Regulation of gene expression and mRNA export by nuclear pore acetylation
3. Mechanisms of rDNA instability and extra-chromosomal rDNA circle accumulation during aging
Funding and partners
- French National Research Agency (ANR), Coordinator of Collaborative Research Project, 2022-2025
- ARC Foundation for Cancer Research, « Programme Labellisé », 2022-2025
- FRM Medical Research Foundation, “Equipe Labellisée”, 2021-2024
Awards and recognitions
European Research Council, Starting Grant (2011-2017)
Publications
-
2018
-
High levels of histones promote whole-genome-duplications and trigger a Swe1WEE1-dependent phosphorylation of Cdc28CDK1
- Douglas Maya Miles
- Xenia Peñate
- Trinidad Sanmartín Olmo
- Frederic Jourquin
- Maria Cruz Muñoz Centeno
- Manuel Mendoza
- Marie-Noelle Simon
- Sebastian Chavez
- Vincent Geli
eLife ; Volume: 7
-
-
2017
-
Cdc14 Localization as a Marker for Mitotic Exit: In Vivo Quantitative Analysis of Cdc14 Release
- Gabriel Neurohr
- Manuel Mendoza
The Mitotic Exit Network ; Volume: 1505 ; Page: 59-67
-
Distinct roles of the polarity factors Boi1 and Boi2 in the control of exocytosis and abscission in budding yeast
- Aina Masgrau
- Andrea Battola
- Trinidad Sanmartin
- Leszek Pryszcz
- Toni Gabaldón
- Manuel Mendoza
Molecular Biology of the Cell ; Volume: 28 ; Page: 3082-3094
-
DNA replication stress: NoCut to the rescue
- Nuno Amaral
- Nicola Brownlow
- Manuel Mendoza
Cell Cycle ; Volume: 16 ; Page: 233-234
-
-
2016
-
The Aurora-B-dependent NoCut checkpoint prevents damage of anaphase bridges after DNA replication stress
- Nuno Amaral
- Alexandre Vendrell
- Charlotta Funaya
- Fatima-Zahra Idrissi
- Michael Maier
- Arun Kumar
- Gabriel Neurohr
- Neus Colomina
- Jordi Torres-Rosell
- María-Isabel Geli
- Manuel Mendoza
Nature Cell Biology ; Volume: 18 ; Page: 516-526
-
Time-Lapse Fluorescence Microscopy of Budding Yeast Cells
- Arun Kumar
- Manuel Mendoza
Yeast Cytokinesis ; Volume: 1369 ; Page: 1-8
-
-
2014
-
Chromosome length and perinuclear attachment constrain resolution of DNA intertwines
- Iris Titos
- Tsvetomira Ivanova
- Manuel Mendoza
Journal of Cell Biology ; Volume: 206 ; Page: 719-733
-
-
2011
-
A Midzone-Based Ruler Adjusts Chromosome Compaction to Anaphase Spindle Length
- Gabriel Neurohr
- Andreas Naegeli
- Iris Titos
- Dominik Theler
- Basil Greber
- Javier Díez
- Toni Gabaldón
- Manuel Mendoza
- Yves Barral
Science ; Volume: 332 ; Page: 465-468
-
-
2009
-
A mechanism for chromosome segregation sensing by the NoCut checkpoint
- Manuel Mendoza
- Caren Norden
- Kathrin Durrer
- Harald Rauter
- Frank Uhlmann
- Yves Barral
Nature Cell Biology ; Volume: 11 ; Page: 477-483
-
-
2008
-
Cytokinesis: Keeping Ring and Membrane Together
- Manuel Mendoza
- Yves Barral
Current Biology ; Volume: 18 ; Page: R479-R480
-
Page 2 of 3

